Grid computing is the use of widely distributed computer resources to reach a common goal. A computing grid can be thought of as a distributed system with non-interactive workloads that involve many files. Grid computing is distinguished from conventional high-performance computing systems such as cluster computing in that grid computers have each node set to perform a different task/application. Grid computers also tend to be more heterogeneous and geographically dispersed than cluster computers. Although a single grid can be dedicated to a particular application, commonly a grid is used for a variety of purposes. Grids are often constructed with general-purpose grid middleware software libraries. Grid sizes can be quite large.
E-Science or eScience is computationally intensive science that is carried out in highly distributed network environments, or science that uses immense data sets that require grid computing; the term sometimes includes technologies that enable distributed collaboration, such as the Access Grid. The term was created by John Taylor, the Director General of the United Kingdom's Office of Science and Technology in 1999 and was used to describe a large funding initiative starting in November 2000. E-science has been more broadly interpreted since then, as "the application of computer technology to the undertaking of modern scientific investigation, including the preparation, experimentation, data collection, results dissemination, and long-term storage and accessibility of all materials generated through the scientific process. These may include data modeling and analysis, electronic/digitized laboratory notebooks, raw and fitted data sets, manuscript production and draft versions, pre-prints, and print and/or electronic publications." In 2014, IEEE eScience Conference Series condensed the definition to "eScience promotes innovation in collaborative, computationally- or data-intensive research across all disciplines, throughout the research lifecycle" in one of the working definitions used by the organizers. E-science encompasses "what is often referred to as big data [which] has revolutionized science... [such as] the Large Hadron Collider (LHC) at CERN... [that] generates around 780 terabytes per year... highly data intensive modern fields of science...that generate large amounts of E-science data include: computational biology, bioinformatics, genomics" and the human digital footprint for the social sciences.
United States federal research funders use the term cyberinfrastructure to describe research environments that support advanced data acquisition, data storage, data management, data integration, data mining, data visualization and other computing and information processing services distributed over the Internet beyond the scope of a single institution. In scientific usage, cyberinfrastructure is a technological and sociological solution to the problem of efficiently connecting laboratories, data, computers, and people with the goal of enabling derivation of novel scientific theories and knowledge.

TeraGrid was an e-Science grid computing infrastructure combining resources at eleven partner sites. The project started in 2001 and operated from 2004 through 2011.

NorduGrid is a collaboration aiming at development, maintenance and support of the free Grid middleware, known as the Advanced Resource Connector (ARC).
The Texas Advanced Computing Center (TACC) at the University of Texas at Austin, United States, is an advanced computing research center that is based on comprehensive advanced computing resources and supports services to researchers in Texas and across the U.S. The mission of TACC is to enable discoveries that advance science and society through the application of advanced computing technologies. Specializing in high-performance computing, scientific visualization, data analysis & storage systems, software, research & development, and portal interfaces, TACC deploys and operates advanced computational infrastructure to enable the research activities of faculty, staff, and students of UT Austin. TACC also provides consulting, technical documentation, and training to support researchers who use these resources. TACC staff members conduct research and development in applications and algorithms, computing systems design/architecture, and programming tools and environments.

nanoHUB.org is a science and engineering gateway comprising community-contributed resources and geared toward education, professional networking, and interactive simulation tools for nanotechnology. Funded by the United States National Science Foundation (NSF), it is a product of the Network for Computational Nanotechnology (NCN). NCN supports research efforts in nanoelectronics; nanomaterials; nanoelectromechanical systems (NEMS); nanofluidics; nanomedicine, nanobiology; and nanophotonics.
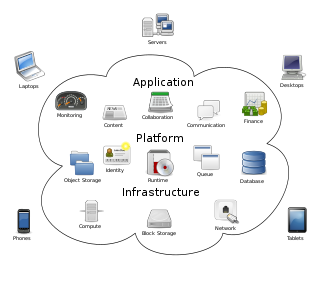
Cloud computing is the on-demand availability of computer system resources, especially data storage and computing power, without direct active management by the user. Large clouds often have functions distributed over multiple locations, each of which is a data center. Cloud computing relies on sharing of resources to achieve coherence and typically uses a pay-as-you-go model, which can help in reducing capital expenses but may also lead to unexpected operating expenses for users.
Techila Distributed Computing Engine is a commercial grid computing software product. It speeds up simulation, analysis and other computational applications by enabling scalability across the IT resources in user's on-premises data center and in the user's own cloud account. Techila Distributed Computing Engine is developed and licensed by Techila Technologies Ltd, a privately held company headquartered in Tampere, Finland. The product is also available as an on-demand solution in Google Cloud Launcher, the online marketplace created and operated by Google. According to IDC, the solution enables organizations to create HPC infrastructure without the major capital investments and operating expenses required by new HPC hardware.

The Laboratory of Parallel and Distributed Systems (LPDS), as a department of MTA SZTAKI, is a research laboratory in distributed grid and cloud technologies. LPDS is a founding member of the Hungarian Grid Competence Centre, the Hungarian National Grid Initiative, and the Hungarian OpenNebula Community, and also coordinates several European grid/cloud projects.

The Grid and Cloud User Support Environment (gUSE), also known as WS-PGRADE /gUSE, is an open source science gateway framework that enables users to access grid and cloud infrastructures. gUSE is developed by the Laboratory of Parallel and Distributed Systems (LPDS) at Institute for Computer Science and Control (SZTAKI) of the Hungarian Academy of Sciences.
A scientific workflow system is a specialized form of a workflow management system designed specifically to compose and execute a series of computational or data manipulation steps, or workflow, in a scientific application.
Integrated computational materials engineering (ICME) involves the integration of experimental results, design models, simulations, and other computational data related to a variety of materials used in multiscale engineering and design. Central to the achievement of ICME goals has been the creation of a cyberinfrastructure, a Web-based, collaborative platform which provides the ability to accumulate, organize and disseminate knowledge pertaining to materials science and engineering to facilitate this information being broadly utilized, enhanced, and expanded.

The iPlant Collaborative, renamed Cyverse in 2017, is a virtual organization created by a cooperative agreement funded by the US National Science Foundation (NSF) to create cyberinfrastructure for the plant sciences (botany). The NSF compared cyberinfrastructure to physical infrastructure, "... the distributed computer, information and communication technologies combined with the personnel and integrating components that provide a long-term platform to empower the modern scientific research endeavor". In September 2013 it was announced that the National Science Foundation had renewed iPlant's funding for a second 5-year term with an expansion of scope to all non-human life science research.
Airavata is an open source software suite that composes, manages, executes, and monitors large-scale applications and workflows on computational resources, ranging from local clusters to national grids, and computing clouds.
HUBzero is an open source software platform for building websites that support scientific activities. The platform allows individuals to create web sites that connect a community in scientific research and educational activities. HUBzero is released under various open source licenses.

The OnlineHPC was a free public web service that supplied tools to deal with high performance computers and online workflow editor. OnlineHPC allowed users to design and execute workflows using the online workflow designer and to work with high performance computers – clusters and clouds. Access to high performance resources was available as directly from the service user interface, as from workflow components. The workflow engine of the OnlineHPC service was Taverna as traditionally used for scientific workflow execution in such domains, as bioinformatics, cheminformatics, medicine, astronomy, social science, music, and digital preservation.
The High-performance Integrated Virtual Environment (HIVE) is a distributed computing environment used for healthcare-IT and biological research, including analysis of Next Generation Sequencing (NGS) data, preclinical, clinical and post market data, adverse events, metagenomic data, etc. Currently it is supported and continuously developed by US Food and Drug Administration, George Washington University, and by DNA-HIVE, WHISE-Global and Embleema. HIVE currently operates fully functionally within the US FDA supporting wide variety (+60) of regulatory research and regulatory review projects as well as for supporting MDEpiNet medical device postmarket registries. Academic deployments of HIVE are used for research activities and publications in NGS analytics, cancer research, microbiome research and in educational programs for students at GWU. Commercial enterprises use HIVE for oncology, microbiology, vaccine manufacturing, gene editing, healthcare-IT, harmonization of real-world data, in preclinical research and clinical studies.
The Open Knowledgebase of Interatomic Models (OpenKIM). is a cyberinfrastructure funded by the United States National Science Foundation (NSF) focused on improving the reliability and reproducibility of molecular and multi-scale simulations in computational materials science. It includes a repository of interatomic potentials that are exhaustively tested with user-developed integrity tests, tools to help select among existing potentials and develop new ones, extensive metadata on potentials and their developers, and standard integration methods for using interatomic potentials in major simulation codes. OpenKIM is a member of DataCite and provides unique DOIs (Digital object identifier) for all archived content on the site (fitted models, validation tests, etc.) in order to properly document and provide recognition to content contributors. OpenKIM is also an eXtreme Science and Engineering Discovery Environment (XSEDE) Science Gateway, and all content on openkim.org is available under open source licenses in support of the open science initiative.
D4Science is an organisation operating a Data Infrastructure offering services by community-driven virtual research environments. In particular, it supports communities of practice willing to implement open science practices. The infrastructure follows the system of systems approach, where the constituent systems offer “resources” assembled together to implement the overall set of D4Science services. In particular, D4Science aggregates “domain agnostic” service providers as well as community-specific ones to build a unifying space where the aggregated resources can be exploited via Virtual research Environments and their services.







