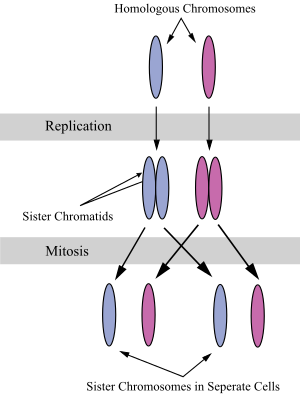
A sister chromatid refers to the identical copies (chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere. In other words, a sister chromatid may also be said to be 'one-half' of the duplicated chromosome. A pair of sister chromatids is called a dyad. A full set of sister chromatids is created during the synthesis (S) phase of interphase, when all the chromosomes in a cell are replicated. The two sister chromatids are separated from each other into two different cells during mitosis or during the second division of meiosis.
Contents
Compare sister chromatids to homologous chromosomes, which are the two different copies of a chromosome that diploid organisms (like humans) inherit, one from each parent. Sister chromatids are by and large identical (since they carry the same alleles, also called variants or versions, of genes) because they derive from one original chromosome. An exception is towards the end of meiosis, after crossing over has occurred, because sections of each sister chromatid may have been exchanged with corresponding sections of the homologous chromatids with which they are paired during meiosis. Homologous chromosomes might or might not be the same as each other because they derive from different parents.
There is evidence that, in some species, sister chromatids are the preferred template for DNA repair. [1] Sister chromatid cohesion is essential for the correct distribution of genetic information between daughter cells and the repair of damaged chromosomes. Defects in this process may lead to aneuploidy and cancer, especially when checkpoints fail to detect DNA damage or when incorrectly attached mitotic spindles do not function properly.
