Related Research Articles

In biochemistry, denaturation is a process in which proteins or nucleic acids lose the quaternary structure, tertiary structure, and secondary structure which is present in their native state, by application of some external stress or compound such as a strong acid or base, a concentrated inorganic salt, an organic solvent, agitation and radiation or heat. If proteins in a living cell are denatured, this results in disruption of cell activity and possibly cell death. Protein denaturation is also a consequence of cell death. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility to aggregation due to the exposure of hydrophobic groups. The loss of solubility as a result of denaturation is called coagulation. Denatured proteins lose their 3D structure and therefore cannot function.

In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the binding site, and residues that catalyse a reaction of that substrate, the catalytic site. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes.

Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology; and it is important in medicine and biotechnology.
Salting out is a purification technique that utilizes the reduced solubility of certain molecules in a solution of very high ionic strength. Salting out is typically used to precipitate large biomolecules, such as proteins or DNA. Because the salt concentration needed for a given protein to precipitate out of the solution differs from protein to protein, a specific salt concentration can be used to precipitate a target protein. This process is also used to concentrate dilute solutions of proteins. Dialysis can be used to remove the salt if needed.

The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in an aqueous solution and exclude water molecules. The word hydrophobic literally means "water-fearing", and it describes the segregation of water and nonpolar substances, which maximizes hydrogen bonding between molecules of water and minimizes the area of contact between water and nonpolar molecules. In terms of thermodynamics, the hydrophobic effect is the free energy change of water surrounding a solute. A positive free energy change of the surrounding solvent indicates hydrophobicity, whereas a negative free energy change implies hydrophilicity.
An artificial membrane, or synthetic membrane, is a synthetically created membrane which is usually intended for separation purposes in laboratory or in industry. Synthetic membranes have been successfully used for small and large-scale industrial processes since the middle of twentieth century. A wide variety of synthetic membranes is known. They can be produced from organic materials such as polymers and liquids, as well as inorganic materials. The most of commercially utilized synthetic membranes in separation industry are made of polymeric structures. They can be classified based on their surface chemistry, bulk structure, morphology, and production method. The chemical and physical properties of synthetic membranes and separated particles as well as a choice of driving force define a particular membrane separation process. The most commonly used driving forces of a membrane process in industry are pressure and concentration gradients. The respective membrane process is therefore known as filtration. Synthetic membranes utilized in a separation process can be of different geometry and of respective flow configuration. They can also be categorized based on their application and separation regime. The best known synthetic membrane separation processes include water purification, reverse osmosis, dehydrogenation of natural gas, removal of cell particles by microfiltration and ultrafiltration, removal of microorganisms from dairy products, and Dialysis.
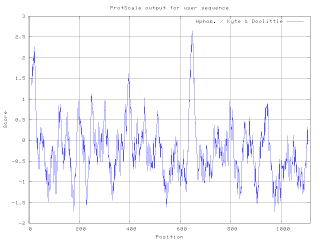
A hydrophilicity plot is a quantitative analysis of the degree of hydrophobicity or hydrophilicity of amino acids of a protein. It is used to characterize or identify possible structure or domains of a protein.
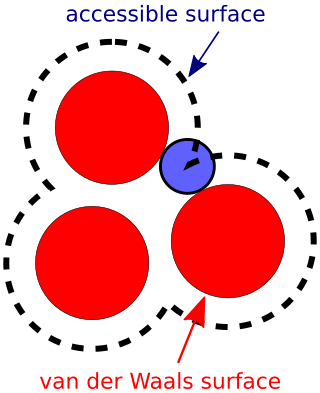
The accessible surface area (ASA) or solvent-accessible surface area (SASA) is the surface area of a biomolecule that is accessible to a solvent. Measurement of ASA is usually described in units of square angstroms. ASA was first described by Lee & Richards in 1971 and is sometimes called the Lee-Richards molecular surface. ASA is typically calculated using the 'rolling ball' algorithm developed by Shrake & Rupley in 1973. This algorithm uses a sphere of a particular radius to 'probe' the surface of the molecule.

Hydrophobic collapse is a proposed process for the production of the 3-D conformation adopted by polypeptides and other molecules in polar solvents. The theory states that the nascent polypeptide forms initial secondary structure creating localized regions of predominantly hydrophobic residues. The polypeptide interacts with water, thus placing thermodynamic pressures on these regions which then aggregate or "collapse" into a tertiary conformation with a hydrophobic core. Incidentally, polar residues interact favourably with water, thus the solvent-facing surface of the peptide is usually composed of predominantly hydrophilic regions.

Half Sphere exposure (HSE) is a protein solvent exposure measure that was first introduced by Hamelryck (2005). Like all solvent exposure measures it measures how buried amino acid residues are in a protein. It is found by counting the number of amino acid neighbors within two half spheres of chosen radius around the amino acid. The calculation of HSE is found by dividing a contact number (CN) sphere in two halves by the plane perpendicular to the Cβ-Cα vector. This simple division of the CN sphere results in two strikingly different measures, HSE-up and HSE-down. HSE-up is defined as the number of Cα atoms in the upper half and analogously HSE-down is defined as the number of Cα atoms in the opposite sphere.
Protein precipitation is widely used in downstream processing of biological products in order to concentrate proteins and purify them from various contaminants. For example, in the biotechnology industry protein precipitation is used to eliminate contaminants commonly contained in blood. The underlying mechanism of precipitation is to alter the solvation potential of the solvent, more specifically, by lowering the solubility of the solute by addition of a reagent.
Stain removal is the process of removing a mark or spot left by one substance on a specific surface like a fabric. A solvent or detergent is generally used to conduct stain removal and many of these are available over the counter.
Hydrophobicity scales are values that define the relative hydrophobicity or hydrophilicity of amino acid residues. The more positive the value, the more hydrophobic are the amino acids located in that region of the protein. These scales are commonly used to predict the transmembrane alpha-helices of membrane proteins. When consecutively measuring amino acids of a protein, changes in value indicate attraction of specific protein regions towards the hydrophobic region inside lipid bilayer.
Self-assembling peptides are a category of peptides which undergo spontaneous assembling into ordered nanostructures. Originally described in 1993, these designer peptides have attracted interest in the field of nanotechnology for their potential for application in areas such as biomedical nanotechnology, tissue cell culturing, molecular electronics, and more.
Residue depth (RD) is a solvent exposure measure that describes to what extent a residue is buried in the protein structure space. It complements the information provided by conventional accessible surface area (ASA).

Peptide amphiphiles (PAs) are peptide-based molecules that self-assemble into supramolecular nanostructures including; spherical micelles, twisted ribbons, and high-aspect-ratio nanofibers. A peptide amphiphile typically comprises a hydrophilic peptide sequence attached to a lipid tail, i.e. a hydrophobic alkyl chain with 10 to 16 carbons. Therefore, they can be considered a type of lipopeptide. A special type of PA, is constituted by alternating charged and neutral residues, in a repeated pattern, such as RADA16-I. The PAs were developed in the 1990s and the early 2000s and could be used in various medical areas including: nanocarriers, nanodrugs, and imaging agents. However, perhaps their main potential is in regenerative medicine to culture and deliver cells and growth factors.
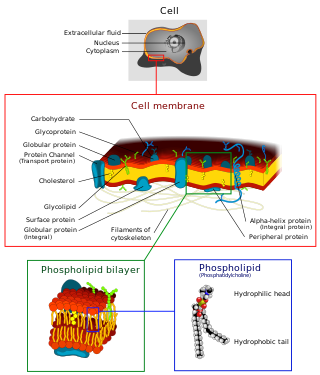
The cell membrane is a biological membrane that separates and protects the interior of a cell from the outside environment. The cell membrane consists of a lipid bilayer, made up of two layers of phospholipids with cholesterols interspersed between them, maintaining appropriate membrane fluidity at various temperatures. The membrane also contains membrane proteins, including integral proteins that span the membrane and serve as membrane transporters, and peripheral proteins that loosely attach to the outer (peripheral) side of the cell membrane, acting as enzymes to facilitate interaction with the cell's environment. Glycolipids embedded in the outer lipid layer serve a similar purpose. The cell membrane controls the movement of substances in and out of a cell, being selectively permeable to ions and organic molecules. In addition, cell membranes are involved in a variety of cellular processes such as cell adhesion, ion conductivity, and cell signalling and serve as the attachment surface for several extracellular structures, including the cell wall and the carbohydrate layer called the glycocalyx, as well as the intracellular network of protein fibers called the cytoskeleton. In the field of synthetic biology, cell membranes can be artificially reassembled.
Computer Atlas of Surface Topography of Proteins (CASTp) aims to provide comprehensive and detailed quantitative characterization of topographic features of protein, is now updated to version 3.0. Since its release in 2006, the CASTp server has ≈45000 visits and fulfills ≈33000 calculation requests annually. CASTp has been proven as a confident tool for a wide range of researches, including investigations of signaling receptors, discoveries of cancer therapeutics, understanding of mechanism of drug actions, studies of immune disorder diseases, analysis of protein–nanoparticle interactions, inference of protein functions and development of high-throughput computational tools. This server is maintained by Jie Liang's lab in University of Illinois at Chicago.
Volume, Area, Dihedral Angle Reporter (VADAR) is a freely available protein structure validation web server that was developed as a collaboration between Dr. Brian Sykes and Dr. David Wishart at the University of Alberta. VADAR consists of over 15 different algorithms and programs for assessing and validating peptide and protein structures from their PDB coordinate data. VADAR is capable of determining secondary structure, identifying and classifying six different types of beta turns, determining and calculating the strength of C=O -- N-H hydrogen bonds, calculating residue-specific accessible surface areas (ASA), calculating residue volumes, determining backbone and side chain torsion angles, assessing local structure quality, evaluating global structure quality, and identifying residue "outliers". The results have been validated through extensive comparison to published data and careful visual inspection. VADAR produces both text and graphical output with most of the quantitative data presented in easily viewed tables. In particular, VADAR's output is presented in a vertical, tabular format with most of the sequence data, residue numbering and any other calculated property or feature presented from top to bottom, rather than from left to right.
Relative accessible surface area or relative solvent accessibility (RSA) of a protein residue is a measure of residue solvent exposure. It can be calculated by formula:
References
Lee B, Richards F. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55:379-400
Greer J, Bush B. (1978) Macromolecular shape and surface maps by solvent exclusion. Proc. Natl. Acad. Sci. USA 75:303-307.
Connolly M. (1983) Solvent-accessible surfaces of proteins and nucleic acids. Science 221:709-713
Chakravarty S, Varadarajan R. (1999) Residue depth: a novel parameter for the analysis of protein structure and stability. Structure Fold. Des. 7:723-732.
Pintar A, Carugo O, Pongor S. (2003) Atom depth in protein structure and function. Trends Biochem. Sci. 28:593-597.
Hamelryck T. (2005) An amino acid has two sides: A new 2D measure provides a different view of solvent exposure. Proteins Struct. Func. Bioinf. 59:38-48.