Staphylococcus aureus is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA), is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.
Pathogenicity islands (PAIs), as termed in 1990, are a distinct class of genomic islands acquired by microorganisms through horizontal gene transfer. Pathogenicity islands are found in both animal and plant pathogens. Additionally, PAIs are found in both gram-positive and gram-negative bacteria. They are transferred through horizontal gene transfer events such as transfer by a plasmid, phage, or conjugative transposon. Therefore, PAIs contribute to microorganisms' ability to evolve.
Virulence factors are cellular structures, molecules and regulatory systems that enable microbial pathogens to achieve the following:

Protein A is a 42 kDa surface protein originally found in the cell wall of the bacteria Staphylococcus aureus. It is encoded by the spa gene and its regulation is controlled by DNA topology, cellular osmolarity, and a two-component system called ArlS-ArlR. It has found use in biochemical research because of its ability to bind immunoglobulins. It is composed of five homologous Ig-binding domains that fold into a three-helix bundle. Each domain is able to bind proteins from many mammalian species, most notably IgGs. It binds the heavy chain within the Fc region of most immunoglobulins and also within the Fab region in the case of the human VH3 family. Through these interactions in serum, where IgG molecules are bound in the wrong orientation, the bacteria disrupts opsonization and phagocytosis.

Hemolysins or haemolysins are lipids and proteins that cause lysis of red blood cells by disrupting the cell membrane. Although the lytic activity of some microbe-derived hemolysins on red blood cells may be of great importance for nutrient acquisition, many hemolysins produced by pathogens do not cause significant destruction of red blood cells during infection. However, hemolysins are often capable of lysing red blood cells in vitro.
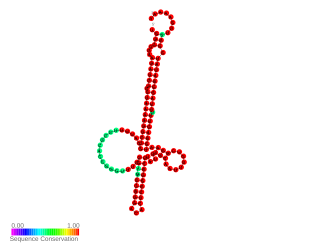
RNAIII is a stable 514 nt regulatory RNA transcribed by the P3 promoter of the Staphylococcus aureus quorum-sensing agr system ). It is the major effector of the agr regulon, which controls the expression of many S. aureus genes encoding exoproteins and cell wall associated proteins plus others encoding regulatory proteins The RNAIII transcript also encodes the 26 amino acid δ-haemolysin peptide (Hld). RNAIII contains many stem loops, most of which match the Shine-Dalgarno sequence involved in translation initiation of the regulated genes. Some of these interactions are inhibitory, others stimulatory; among the former is the regulatory protein Rot. In vitro, RNAIII is expressed post exponentially, inhibiting translation of the surface proteins, notably protein A, while stimulating that of the exoproteins, many of which are tissue-degrading enzymes or cytolysins. Among the latter is the important virulence factor, α-hemolysin (Hla), whose translation RNAIII activates by preventing the formation of an inhibitory foldback loop in the hla mRNA leader.
'Staphylococcus aureus delta toxin is a toxin produced by Staphylococcus aureus. It has a wide spectrum of cytolytic activity.
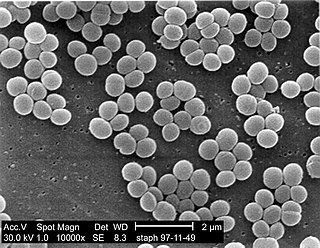
Staphylococcus is a genus of Gram-positive bacteria in the family Staphylococcaceae from the order Bacillales. Under the microscope, they appear spherical (cocci), and form in grape-like clusters. Staphylococcus species are facultative anaerobic organisms.
Bacterial small RNAs (bsRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).
SaPIs are a family of ~15 kb mobile genetic elements resident in the genomes of the vast majority of S. aureus strains. Much like bacteriophages, SaPIs can be transferred to uninfected cells and integrate into the host chromosome. Unlike the bacterial viruses, however, integrated SaPIs are mobilized by host infection with "helper" bacteriophages. SaPIs are used by the host bacteria to co-opt the phage reproduction cycle for their own genetic transduction and also inhibit phage reproduction in the process.
In molecular biology, the domain B, refers to the immunoglobulin-binding domain found in the Staphylococcus aureus virulence factor protein A (SpA). Hence, it is abbreviated to SpAB.

Glutamyl endopeptidase is an extracellular bacterial serine protease of the glutamyl endopeptidase I family that was initially isolated from the Staphylococcus aureus strain V8. The protease is, hence, commonly referred to as "V8 protease", or alternatively SspA from its corresponding gene.

Aureolysin is an extracellular metalloprotease expressed by Staphylococcus aureus. This protease is a major contributor to the bacterium's virulence, or ability to cause disease, by cleaving host factors of the innate immune system as well as regulating S. aureus secreted toxins and cell wall proteins. To catalyze its enzymatic activities, aureolysin requires zinc and calcium which it obtains from the extracellular environment within the host.
Staphylococcus pseudintermedius is a gram positive coccus bacteria of the genus Staphylococcus found worldwide. It is primarily a pathogen for domestic animals, but has been known to affect humans as well. S. pseudintermedius is an opportunistic pathogen that secretes immune modulating virulence factors, has many adhesion factors, and the potential to create biofilms, all of which help to determine the pathogenicity of the bacterium. Diagnoses of Staphylococcus pseudintermedius have traditionally been made using cytology, plating, and biochemical tests. More recently, molecular technologies like MALDI-TOF, DNA hybridization and PCR have become preferred over biochemical tests for their more rapid and accurate identifications. This includes the identification and diagnosis of antibiotic resistant strains.
In molecular biology the small pathogenicity island RNA X gene is a bacterial non-coding RNA. It was discovered in a large-scale analysis of Staphylococcus aureus. SprX was shown to influence antibiotic resistance of the bacteria to Vancomycin and Teicoplanin glycopeptides, which are used to treat MRSA infections. In this study the authors identified a SprX target, stage V sporulation protein G. By reducing Spo VG expression levels, SprX affects S. aureus resistance to the glycopeptide antibiotics. Further work demonstrated its involvement in the regulation of pathogenicity factors.
Although cell wall carbohydrates are ideal immunotherapeutic targets due to their abundance in bacteria and high level of conservation, their poor immunogenicity compared with protein targets complicates their use for the development of protective antibodies. A lysibody is a chimeric antibody in which the Fab region is the binding domain from a bacteriophage lysin, or the binding domain from an autolysin or bacteriocin, all of which bind to bacterial cell wall carbohydrate epitopes. This is linked to the Fc of Immunoglobulin G (IgG). The chimera forms a stable homodimer held together by hinge-region disulfide bonds. Thus, lysibodies are homodimeric hybrid immunoglobulin G molecules that can bind with high affinity and specificity to a carbohydrate substrate in the bacterial cell wall peptidoglycan. Lysibodies behave like authentic IgG by binding at high affinity to their bacterial wall receptor, fix complement and therefore promote phagocytosis by macrophages and neutrophils, protecting mice from infection in model systems. Since cell wall hydrolases, autolysins and bacteriocins are ubiquitous in nature, production of lysibodies specific for difficult to treat pathogenic bacteria is possible.
RNA thermometers (RNATs) regulate gene expression in response to temperature, allowing pathogens such as Neisseria meningitidis to switch on silent genes after entering the host organism. However the temperature for expression of Neisseria virulence-associated traits is 42 °C while other bacterial pathogen RNATs require 37 °C. This is probably because N. meningitidis is an obligate commensal of the human nasopharynx and becomes pathogenic during inflammation due to viral infection. Three independent RNA thermosensors were identified in the 5′UTRs of genes needed for: capsule biosynthesis (cssA), the expression of factor H binding protein (fHbp) and sialylation of lipopolysaccharide, which is essential for bacterial resistance against immune killing (lst). The very different nucleotide sequence and predicted inhibitory structures of the three RNATs indicate that they have evolved independently.
The SprA1/SPrA1as toxin/antitoxin system identified in Staphylococcus aureus, belongs to the Type I system encoding toxin protein: SprA1 and antitoxin RNA: SprA1as. The SprA1as postranscriptionally regulates SprA1 encoding small membrane damaging protein PepA1.
Accessory gene regulator (agr) is a complex 5 gene locus that is a global regulator of virulence in Staphylococcus aureus. It encodes a two-component transcriptional quorum-sensing (QS) system activated by an autoinducing, thiolactone-containing cyclic peptide (AIP).