An antibody (Ab) is the secreted form of a B cell receptor; the term immunoglobulin can refer to either the membrane-bound form or the secreted form of the B cell receptor, but they are, broadly speaking, the same protein, and so the terms are often treated as synonymous. Antibodies are large, Y-shaped proteins belonging to the immunoglobulin superfamily which are used by the immune system to identify and neutralize foreign objects such as bacteria and viruses, including those that cause disease. Antibodies can recognize virtually any size antigen with diverse chemical compositions from molecules. Each antibody recognizes one or more specific antigens. This term literally means "antibody generator", as it is the presence of an antigen that drives the formation of an antigen-specific antibody. Each tip of the "Y" of an antibody contains a paratope that specifically binds to one particular epitope on an antigen, allowing the two molecules to bind together with precision. Using this mechanism, antibodies can effectively "tag" a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly.
CD32, also known as FcγRII or FCGR2, is a surface receptor glycoprotein belonging to the Ig gene superfamily. CD32 can be found on the surface of a variety of immune cells. CD32 has a low-affinity for the Fc region of IgG antibodies in monomeric form, but high affinity for IgG immune complexes. CD32 has two major functions: cellular response regulation, and the uptake of immune complexes. Cellular responses regulated by CD32 include phagocytosis, cytokine stimulation, and endocytic transport. Dysregulated CD32 is associated with different forms of autoimmunity, including systemic lupus erythematosus. In humans, there are three major CD32 subtypes: CD32A, CD32B, and CD32C. While CD32A and CD32C are involved in activating cellular responses, CD32B is inhibitory.

A monoclonal antibody is an antibody produced from a cell lineage made by cloning a unique white blood cell. All subsequent antibodies derived this way trace back to a unique parent cell.

Superantigens (SAgs) are a class of antigens that result in excessive activation of the immune system. Specifically they cause non-specific activation of T-cells resulting in polyclonal T cell activation and massive cytokine release. Superantigens act by binding to the MHC proteins on antigen-presenting cells (APCs) and to the TCRs on their adjacent helper T-cells, bringing the signaling molecules together, and thus leading to the activation of the T-cells, regardless of the peptide displayed on the MHC molecule. SAgs are produced by some pathogenic viruses and bacteria most likely as a defense mechanism against the immune system. Compared to a normal antigen-induced T-cell response where 0.0001-0.001% of the body's T-cells are activated, these SAgs are capable of activating up to 20% of the body's T-cells. Furthermore, Anti-CD3 and Anti-CD28 antibodies (CD28-SuperMAB) have also shown to be highly potent superantigens.

The classical complement pathway is one of three pathways which activate the complement system, which is part of the immune system. The classical complement pathway is initiated by antigen-antibody complexes with the antibody isotypes IgG and IgM.
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation, compared to other chromatographic methods.
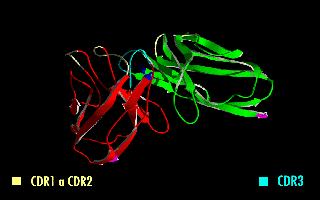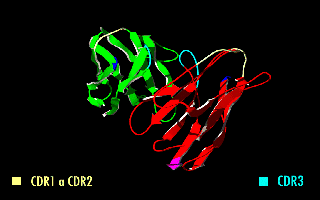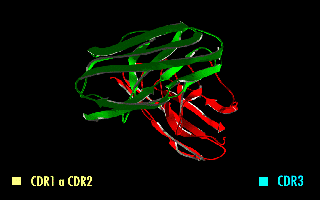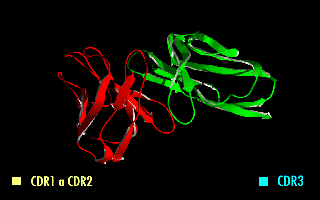
A single-chain variable fragment (scFv) is not actually a fragment of an antibody, but instead is a fusion protein of the variable regions of the heavy (VH) and light chains (VL) of immunoglobulins, connected with a short linker peptide of ten to about 25 amino acids. The linker is usually rich in glycine for flexibility, as well as serine or threonine for solubility, and can either connect the N-terminus of the VH with the C-terminus of the VL, or vice versa. This protein retains the specificity of the original immunoglobulin, despite removal of the constant regions and the introduction of the linker. The image to the right shows how this modification usually leaves the specificity unaltered.

Antibody-dependent cellular cytotoxicity (ADCC), also referred to as antibody-dependent cell-mediated cytotoxicity, is a mechanism of cell-mediated immune defense whereby an effector cell of the immune system kills a target cell, whose membrane-surface antigens have been bound by specific antibodies. It is one of the mechanisms through which antibodies, as part of the humoral immune response, can act to limit and contain infection.
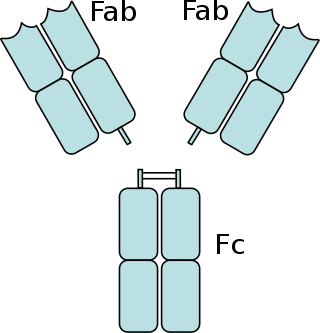
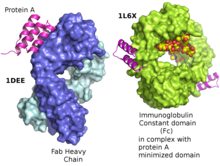
The fragment crystallizable region is the tail region of an antibody that interacts with cell surface receptors called Fc receptors and some proteins of the complement system. This region allows antibodies to activate the immune system, for example, through binding to Fc receptors. In IgG, IgA and IgD antibody isotypes, the Fc region is composed of two identical protein fragments, derived from the second and third constant domains of the antibody's two heavy chains; IgM and IgE Fc regions contain three heavy chain constant domains in each polypeptide chain. The Fc regions of IgGs bear a highly conserved N-glycosylation site. Glycosylation of the Fc fragment is essential for Fc receptor-mediated activity. The N-glycans attached to this site are predominantly core-fucosylated diantennary structures of the complex type. In addition, small amounts of these N-glycans also bear bisecting GlcNAc and α-2,6 linked sialic acid residues.
CD22, or cluster of differentiation-22, is a molecule belonging to the SIGLEC family of lectins. It is found on the surface of mature B cells and to a lesser extent on some immature B cells. Generally speaking, CD22 is a regulatory molecule that prevents the overactivation of the immune system and the development of autoimmune diseases.
MSCRAMM adhesin proteins mediate the initial attachment of bacteria to host tissue, providing a critical step to establish infection.

Hemolysins or haemolysins are lipids and proteins that cause lysis of red blood cells by disrupting the cell membrane. Although the lytic activity of some microbe-derived hemolysins on red blood cells may be of great importance for nutrient acquisition, many hemolysins produced by pathogens do not cause significant destruction of red blood cells during infection. However, hemolysins are often capable of lysing red blood cells in vitro.

In immunology, antibodies are classified into several types called isotypes or classes. The variable (V) regions near the tip of the antibody can differ from molecule to molecule in countless ways, allowing it to specifically target an antigen . In contrast, the constant (C) regions only occur in a few variants, which define the antibody's class. Antibodies of different classes activate distinct effector mechanisms in response to an antigen . They appear at different stages of an immune response, differ in structural features, and in their location around the body.
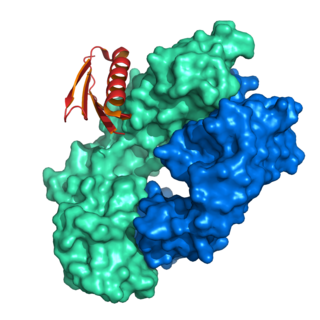
Protein L was first isolated from the surface of bacterial species Peptostreptococcus magnus and was found to bind immunoglobulins through L chain interaction, from which the name was suggested. It consists of 719 amino acid residues. The molecular weight of protein L purified from the cell walls of Peptostreptoccus magnus was first estimated as 95kD by SDS-PAGE in the presence of reducing agent 2-mercaptoethanol, while the molecular weight was determined to 76kD by gel chromatography in the presence of 6 M guanidine HCl. Protein L does not contain any interchain disulfide loops, nor does it consist of disulfide-linked subunits. It is an acidic molecule with a pI of 4.0. Unlike protein A and protein G, which bind to the Fc region of immunoglobulins (antibodies), protein L binds antibodies through light chain interactions. Since no part of the heavy chain is involved in the binding interaction, Protein L binds a wider range of antibody classes than protein A or G. Protein L binds to representatives of all antibody classes, including IgG, IgM, IgA, IgE and IgD. Single chain variable fragments (scFv) and Fab fragments also bind to protein L.
Protein A/G is a recombinant fusion protein that combines IgG binding domains of both protein A and protein G. Protein A/G contains four Fc binding domains from protein A and two from protein G, yielding a final mass of 50,460 daltons. The binding of protein A/G is less pH-dependent than protein A, but otherwise has the additive properties of protein A and G.
Small modular immunopharmaceuticals, or SMIPs for short, are artificial proteins that are intended for use as pharmaceutical drugs. They are largely built from parts of antibodies (immunoglobulins), and like them have a binding site for antigens that could be used for monoclonal antibody therapy. SMIPs have similar biological half-life and, being smaller than antibodies, are reasoned to have better tissue penetration properties. They were invented by Trubion and are now being developed by Emergent BioSolutions, which acquired Trubion in 2010.
A bispecific monoclonal antibody is an artificial protein that can simultaneously bind to two different types of antigen or two different epitopes on the same antigen. Naturally occurring antibodies typically only target one antigen. BsAbs can be manufactured in several structural formats. BsAbs can be designed to recruit and activate immune cells, to interfere with receptor signaling and inactivate signaling ligands, and to force association of protein complexes. BsAbs have been explored for cancer immunotherapy, drug delivery, and Alzeimer's disease.

Affitins are artificial proteins with the ability to selectively bind antigens. They are structurally derived from the DNA binding protein Sac7d, found in Sulfolobus acidocaldarius, a microorganism belonging to the archaeal domain. By randomizing the amino acids on the binding surface of Sac7d and subjecting the resulting protein library to rounds of ribosome display, the affinity can be directed towards various targets, such as peptides, proteins, viruses, and bacteria.
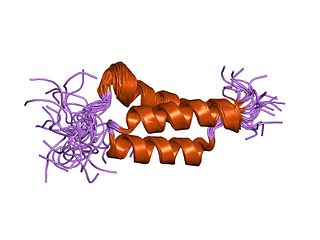
In molecular biology, the domain B, refers to the immunoglobulin-binding domain found in the Staphylococcus aureus virulence factor protein A (SpA). Hence, it is abbreviated to SpAB.
Recombinant antibodies are antibody fragments produced by using recombinant antibody coding genes. They mostly consist of a heavy and light chain of the variable region of immunoglobulin. Recombinant antibodies have many advantages in both medical and research applications, which make them a popular subject of exploration and new production against specific targets. The most commonly used form is the single chain variable fragment (scFv), which has shown the most promising traits exploitable in human medicine and research. In contrast to monoclonal antibodies produced by hybridoma technology, which may lose the capacity to produce the desired antibody over time or the antibody may undergo unwanted changes, which affect its functionality, recombinant antibodies produced in phage display maintain high standard of specificity and low immunogenicity.