fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.

The 245 nucleotide sRNA of Escherichia coli, CsrC, was discovered using a genetic screen for factors that regulate glycogen biosynthesis. CsrC RNA binds multiple copies of CsrA, a protein that post-transcriptionally regulates central carbon flux, biofilm formation and motility in E. coli. CsrC antagonises the regulatory effects of CsrA, presumably by sequestering this protein. The discovery of CsrC is intriguing, in that a similar sRNA, CsrB, performs essentially the same function. Both sRNAs possess similar imperfect repeat sequences, primarily localised in the loops of predicted hairpins, which may serve as CsrA binding elements. Transcription of csrC increases as the culture approaches the stationary phase of growth and is indirectly activated by CsrA via the response regulator UvrY [1]. This RNA was also discovered in E. coli during a large scale screen [2]. The gene called SraK, was highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well [2].

The gcvB RNA gene encodes a small non-coding RNA involved in the regulation of a number of amino acid transport systems as well as amino acid biosynthetic genes. The GcvB gene is found in enteric bacteria such as Escherichia coli. GcvB regulates genes by acting as an antisense binding partner of the mRNAs for each regulated gene. This binding is dependent on binding to a protein called Hfq. Transcription of the GcvB RNA is activated by the adjacent GcvA gene and repressed by the GcvR gene. A deletion of GcvB RNA from Y. pestis changed colony shape as well as reducing growth. It has been shown by gene deletion that GcvB is a regulator of acid resistance in E. coli. GcvB enhances the ability of the bacterium to survive low pH by upregulating the levels of the alternate sigma factor RpoS. A polymeric form of GcvB has recently been identified. Interaction of GcvB with small RNA SroC triggers the degradation of GcvB by RNase E, lifting the GcvB-mediated mRNA repression of its target genes.

The SraC/RyeA RNA is a non-coding RNA that was discovered in E. coli during two large scale screens for RNAs. The function of this RNA is currently unknown. This RNA overlaps the SdsR/RyeB RNA on the opposite strand suggesting that the two RNAs may act in a concerted manner.

The OmrA-B RNA gene family is a pair of homologous OmpR-regulated small non-coding RNA that was discovered in E. coli during two large-scale screens. OmrA-B is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well. RygB is adjacent to RygA a closely related RNA. These RNAs bind to the Hfq protein and regulate gene expression by antisense binding. They negatively regulate the expression of several genes encoding outer membrane proteins, including cirA, CsgD, fecA, fepA and ompT by binding in the vicinity of the Shine-Dalgarno sequence, suggesting the control of these targets is dependent on Hfq protein and RNase E. Taken together, these data suggest that OmrA-B participates in the regulation of outer membrane composition, responding to environmental conditions.
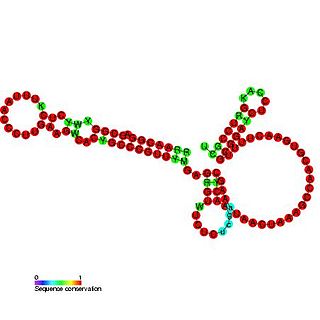
OxyS RNA is a small non-coding RNA which is induced in response to oxidative stress in Escherichia coli. This RNA acts as a global regulator to activate or repress the expression of as many as 40 genes, by an antisense mechanism, including the fhlA-encoded transcriptional activator and the rpoS-encoded sigma(s) subunit of RNA polymerase. OxyS is bound by the Hfq protein, that increases the OxyS RNA interaction with its target messages. Binding to Hfq alters the conformation of OxyS. The 109 nucleotide RNA is thought to be composed of three stem-loops.
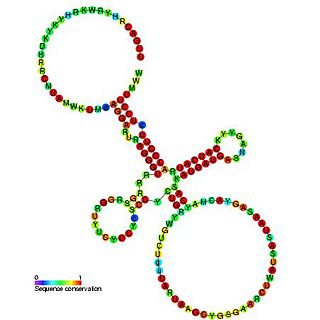
Sib RNA refers to a group of related non-coding RNA. They were originally named QUAD RNA after they were discovered as four repeat elements in Escherichia coli intergenic regions. The family was later renamed Sib when it was discovered that the number of repeats is variable in other species and in other E. coli strains.
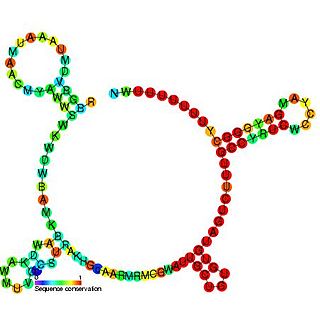
The RprA RNA gene encodes a 106 nucleotide regulatory non-coding RNA. Translational regulation of the stationary phase sigma factor RpoS is mediated by the formation of a double-stranded RNA stem-loop structure in the upstream region of the rpoS messenger RNA, occluding the translation initiation site. Clones carrying rprA increased the translation of RpoS. As with DsrA, RprA is predicted to form three stem-loops. Thus, at least two small RNAs, DsrA and RprA, participate in the positive regulation of RpoS translation. RprA also appears to bind to the RpoS leader. RprA is non-essential. Wasserman et al. demonstrated that this RNA is bound by the Hfq protein. Binding to Hfq alters the conformation of RprA. In the presence of Hfq the stability of RprA is influenced by the osmolarity of the cell, this is dependent on the endoribonuclease RNase E.

RyhB RNA is a 90 nucleotide RNA that down-regulates a set of iron-storage and iron-using proteins when iron is limiting; it is itself negatively regulated by the ferric uptake repressor protein, Fur.

Spot 42 RNA is a regulatory non-coding bacterial small RNA encoded by the spf gene. Spf is found in gammaproteobacteria and the majority of experimental work on Spot42 has been performed in Escherichia coli and recently in Aliivibrio salmonicida. In the cell Spot42 plays essential roles as a regulator in carbohydrate metabolism and uptake, and its expression is activated by glucose, and inhibited by the cAMP-CRP complex.

The MicA RNA is a small non-coding RNA that was discovered in E. coli during a large scale screen. Expression of SraD is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well.

In molecular biology the ArcZ RNA is a small non-coding RNA (ncRNA). It is the functional product of a gene which is not translated into protein. ArcZ is an Hfq binding RNA that functions as an antisense regulator of a number of protein coding genes.
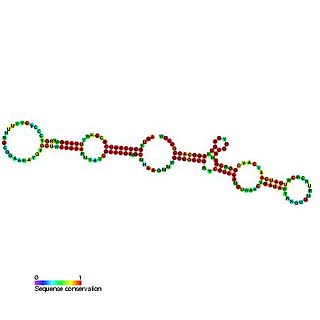
The GlmY RNA family consists of a number of bacterial RNA genes of around 167 bases in length. The GlmY RNA gene is present in Escherichia coli, Shigella flexneri, Yersinia pestis and Salmonella species, where it is found between the yfhK and purL genes. It was originally predicted in a bioinformatic screen for novel ncRNAs in E. coli.
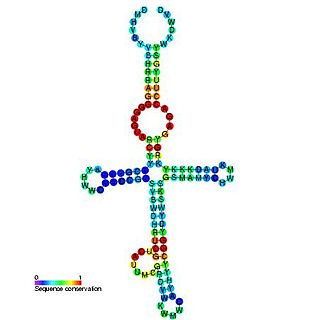
The yybP-ykoY leader RNA element was originally discovered in E. coli during a large scale screen and was named SraF. This family was later found to exist upstream of related families of protein genes in many bacteria, including the yybP and ykoY genes in B. subtilis. The specific functions of these proteins are unknown, but this structured RNA element may be involved in their genetic regulation as a riboswitch. The yybP-ykoY element was later proposed to be manganese-responsive after another associated family of genes, YebN/MntP, was shown to encode Mn2+ efflux pumps in several bacteria. Genetic data and a crystal structure confirmed that yybp-ykoY is a manganese riboswitch that directly binds Mn2+
In a screen of the Bacillus subtilis genome for genes encoding ncRNAs, Saito et al. focused on 123 intergenic regions (IGRs) over 500 base pairs in length, the authors analyzed expression from these regions. Seven IGRs termed bsrC, bsrD, bsrE, bsrF, bsrG, bsrH and bsrI expressed RNAs smaller than 380 nt. All the small RNAs except BsrD RNA were expressed in transformed Escherichia coli cells harboring a plasmid with PCR-amplified IGRs of B. subtilis, indicating that their own promoters independently express small RNAs. Under non-stressed condition, depletion of the genes for the small RNAs did not affect growth. Although their functions are unknown, gene expression profiles at several time points showed that most of the genes except for bsrD were expressed during the vegetative phase, but undetectable during the stationary phase. Mapping the 5' ends of the 6 small RNAs revealed that the genes for BsrE, BsrF, BsrG, BsrH, and BsrI RNAs are preceded by a recognition site for RNA polymerase sigma factor σA.

Mycobacterium tuberculosis contains at least nine small RNA families in its genome. The small RNA (sRNA) families were identified through RNomics – the direct analysis of RNA molecules isolated from cultures of Mycobacterium tuberculosis. The sRNAs were characterised through RACE mapping and Northern blot experiments. Secondary structures of the sRNAs were predicted using Mfold.
Escherichia coli contains a number of small RNAs located in intergenic regions of its genome. The presence of at least 55 of these has been verified experimentally. 275 potential sRNA-encoding loci were identified computationally using the QRNA program. These loci will include false positives, so the number of sRNA genes in E. coli is likely to be less than 275. A computational screen based on promoter sequences recognised by the sigma factor sigma 70 and on Rho-independent terminators predicted 24 putative sRNA genes, 14 of these were verified experimentally by northern blotting. The experimentally verified sRNAs included the well characterised sRNAs RprA and RyhB. Many of the sRNAs identified in this screen, including RprA, RyhB, SraB and SraL, are only expressed in the stationary phase of bacterial cell growth. A screen for sRNA genes based on homology to Salmonella and Klebsiella identified 59 candidate sRNA genes. From this set of candidate genes, microarray analysis and northern blotting confirmed the existence of 17 previously undescribed sRNAs, many of which bind to the chaperone protein Hfq and regulate the translation of RpoS. UptR sRNA transcribed from the uptR gene is implicated in suppressing extracytoplasmic toxicity by reducing the amount of membrane-bound toxic hybrid protein.
The SraL RNA, also known as RyjA, is a small non-coding RNA discovered in E. coli, and later in Salmonella Tiphimurium. This ncRNA was found to be expressed only in stationary phase. It may possibly play a role in Salmonella virulence. The major stationary phase regulator RpoS is transcriptionally regulating SraL and directly binds to the sraL gene promoter. SraL down-regulates the expression of the ribosome-associated chaperone Trigger Factor (TF), which is involved in the folding of the newly synthesised cystolic proteins.





















