Related Research Articles
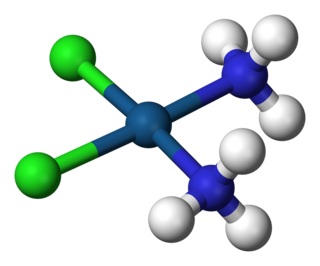
A coordination complex consists of a central atom or ion, which is usually metallic and is called the coordination centre, and a surrounding array of bound molecules or ions, that are in turn known as ligands or complexing agents. Many metal-containing compounds, especially those that include transition metals, are coordination complexes.
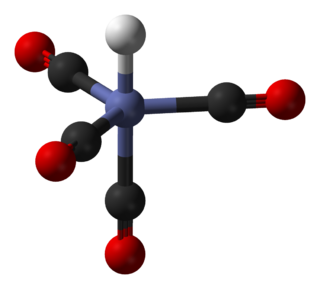
In coordination chemistry, a ligand is an ion or molecule that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's electron pairs, often through Lewis bases. The nature of metal–ligand bonding can range from covalent to ionic. Furthermore, the metal–ligand bond order can range from one to three. Ligands are viewed as Lewis bases, although rare cases are known to involve Lewis acidic "ligands".

In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may include other proteins, enzyme substrates, second messengers, hormones, or allosteric modulators. The binding event is often, but not always, accompanied by a conformational change that alters the protein's function. Binding to protein binding sites is most often reversible, but can also be covalent reversible or irreversible.

In biochemistry and pharmacology, receptors are chemical structures, composed of protein, that receive and transduce signals that may be integrated into biological systems. These signals are typically chemical messengers which bind to a receptor and cause some form of cellular/tissue response, e.g. a change in the electrical activity of a cell. There are three main ways the action of the receptor can be classified: relay of signal, amplification, or integration. Relaying sends the signal onward, amplification increases the effect of a single ligand, and integration allows the signal to be incorporated into another biochemical pathway.

Drug design, often referred to as rational drug design or simply rational design, is the inventive process of finding new medications based on the knowledge of a biological target. The drug is most commonly an organic small molecule that activates or inhibits the function of a biomolecule such as a protein, which in turn results in a therapeutic benefit to the patient. In the most basic sense, drug design involves the design of molecules that are complementary in shape and charge to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on computer modeling techniques. This type of modeling is sometimes referred to as computer-aided drug design. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as structure-based drug design. In addition to small molecules, biopharmaceuticals including peptides and especially therapeutic antibodies are an increasingly important class of drugs and computational methods for improving the affinity, selectivity, and stability of these protein-based therapeutics have also been developed.
Crystal field theory (CFT) describes the breaking of degeneracies of electron orbital states, usually d or f orbitals, due to a static electric field produced by a surrounding charge distribution. This theory has been used to describe various spectroscopies of transition metal coordination complexes, in particular optical spectra (colors). CFT successfully accounts for some magnetic properties, colors, hydration enthalpies, and spinel structures of transition metal complexes, but it does not attempt to describe bonding. CFT was developed by physicists Hans Bethe and John Hasbrouck van Vleck in the 1930s. CFT was subsequently combined with molecular orbital theory to form the more realistic and complex ligand field theory (LFT), which delivers insight into the process of chemical bonding in transition metal complexes.
Ligand field theory (LFT) describes the bonding, orbital arrangement, and other characteristics of coordination complexes. It represents an application of molecular orbital theory to transition metal complexes. A transition metal ion has nine valence atomic orbitals - consisting of five nd, one (n+1)s, and three (n+1)p orbitals. These orbitals are of appropriate energy to form bonding interaction with ligands. The LFT analysis is highly dependent on the geometry of the complex, but most explanations begin by describing octahedral complexes, where six ligands coordinate to the metal. Other complexes can be described by reference to crystal field theory.

Ligand-gated ion channels (LICs, LGIC), also commonly referred to as ionotropic receptors, are a group of transmembrane ion-channel proteins which open to allow ions such as Na+, K+, Ca2+, and/or Cl− to pass through the membrane in response to the binding of a chemical messenger (i.e. a ligand), such as a neurotransmitter.

Fas ligand is a type-II transmembrane protein that belongs to the tumor necrosis factor (TNF) family. Its binding with its receptor induces apoptosis. Fas ligand/receptor interactions play an important role in the regulation of the immune system and the progression of cancer.

In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ligare, which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure.

In chemistry, octahedral molecular geometry, also called square bipyramidal, describes the shape of compounds with six atoms or groups of atoms or ligands symmetrically arranged around a central atom, defining the vertices of an octahedron. The octahedron has eight faces, hence the prefix octa. The octahedron is one of the Platonic solids, although octahedral molecules typically have an atom in their centre and no bonds between the ligand atoms. A perfect octahedron belongs to the point group Oh. Examples of octahedral compounds are sulfur hexafluoride SF6 and molybdenum hexacarbonyl Mo(CO)6. The term "octahedral" is used somewhat loosely by chemists, focusing on the geometry of the bonds to the central atom and not considering differences among the ligands themselves. For example, [Co(NH3)6]3+, which is not octahedral in the mathematical sense due to the orientation of the N−H bonds, is referred to as octahedral.
In biochemistry, an orphan receptor is a protein that has a similar structure to other identified receptors but whose endogenous ligand has not yet been identified. If a ligand for an orphan receptor is later discovered, the receptor is referred to as an "adopted orphan". Conversely, the term orphan ligand refers to a biological ligand whose cognate receptor has not yet been identified.

In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules using, for example, scoring functions.

In coordination chemistry, a bridging ligand is a ligand that connects two or more atoms, usually metal ions. The ligand may be atomic or polyatomic. Virtually all complex organic compounds can serve as bridging ligands, so the term is usually restricted to small ligands such as pseudohalides or to ligands that are specifically designed to link two metals.
The retinoid X receptor (RXR) is a type of nuclear receptor that is activated by 9-cis retinoic acid, which is discussed controversially to be of endogenous relevance, and 9-cis-13,14-dihydroretinoic acid, which is likely to be the major endogenous mammalian RXR-selective agonist.
In the field of molecular biology, nuclear receptors are a class of proteins responsible for sensing steroids, thyroid hormones, vitamins, and certain other molecules. These receptors work with other proteins to regulate the expression of specific genes thereby controlling the development, homeostasis, and metabolism of the organism.

In coordination chemistry, denticity refers to the number of donor groups in a given ligand that bind to the central metal atom in a coordination complex. In many cases, only one atom in the ligand binds to the metal, so the denticity equals one, and the ligand is said to be monodentate. Ligands with more than one bonded atom are called polydentate or multidentate. The denticity of a ligand is described with the Greek letter κ ('kappa'). For example, κ6-EDTA describes an EDTA ligand that coordinates through 6 non-contiguous atoms.
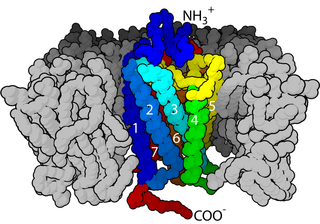
Cell surface receptors are receptors that are embedded in the plasma membrane of cells. They act in cell signaling by receiving extracellular molecules. They are specialized integral membrane proteins that allow communication between the cell and the extracellular space. The extracellular molecules may be hormones, neurotransmitters, cytokines, growth factors, cell adhesion molecules, or nutrients; they react with the receptor to induce changes in the metabolism and activity of a cell. In the process of signal transduction, ligand binding affects a cascading chemical change through the cell membrane.
The IUPHAR/BPS Guide to PHARMACOLOGY is an open-access website, acting as a portal to information on the biological targets of licensed drugs and other small molecules. The Guide to PHARMACOLOGY is developed as a joint venture between the International Union of Basic and Clinical Pharmacology (IUPHAR) and the British Pharmacological Society (BPS). This replaces and expands upon the original 2009 IUPHAR Database. The Guide to PHARMACOLOGY aims to provide a concise overview of all pharmacological targets, accessible to all members of the scientific and clinical communities and the interested public, with links to details on a selected set of targets. The information featured includes pharmacological data, target, and gene nomenclature, as well as curated chemical information for ligands. Overviews and commentaries on each target family are included, with links to key references.
An enzyme modulator is a type of drug which modulates enzymes. They include enzyme inhibitors and enzyme inducers. In an homogeneous assay, "an enzyme modulator ... is covalently linked to the ligand which competes with free ligand from the test sample for the available antibodies."
References
- ↑ Shriver and Atkins. Inorganic Chemistry (5th Edition). W. H. Freeman and Company, New York, 2010, pp 201.