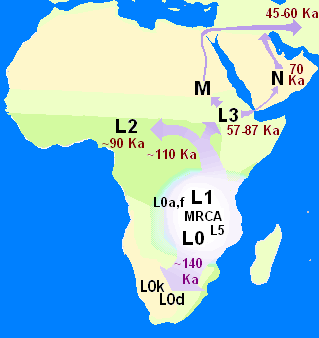
In human genetics, the Mitochondrial Eve is the matrilineal most recent common ancestor (MRCA) of all living humans. In other words, she is defined as the most recent woman from whom all living humans descend in an unbroken line purely through their mothers and through the mothers of those mothers, back until all lines converge on one woman.
In human genetics, the Y-chromosomal most recent common ancestor is the patrilineal most recent common ancestor (MRCA) from whom all currently living humans are descended. He is the most recent male from whom all living humans are descended through an unbroken line of their male ancestors. The term Y-MRCA reflects the fact that the Y chromosomes of all currently living human males are directly derived from the Y chromosome of this remote ancestor. The analogous concept of the matrilineal most recent common ancestor is known as "Mitochondrial Eve", the most recent woman from whom all living humans are descended matrilineally. As with "Mitochondrial Eve", the title of "Y-chromosomal Adam" is not permanently fixed to a single individual, but can advance over the course of human history as paternal lineages become extinct.
Y-chromosomal Aaron is the name given to the hypothesized most recent common ancestor of the patrilineal Jewish priestly caste known as Kohanim. According to the traditional understanding of the Hebrew Bible, this ancestor was Aaron, the brother of Moses.

A haplotype is a group of alleles in an organism that are inherited together from a single parent.
In biology and genetic genealogy, the most recent common ancestor (MRCA), also known as the last common ancestor (LCA), of a set of organisms is the most recent individual from which all the organisms of the set are descended. The term is also used in reference to the ancestry of groups of genes (haplotypes) rather than organisms.
Genetic genealogy is the use of genealogical DNA tests, i.e., DNA profiling and DNA testing, in combination with traditional genealogical methods, to infer genetic relationships between individuals. This application of genetics came to be used by family historians in the 21st century, as DNA tests became affordable. The tests have been promoted by amateur groups, such as surname study groups or regional genealogical groups, as well as research projects such as the Genographic Project.
A genealogical DNA test is a DNA-based genetic test used in genetic genealogy that looks at specific locations of a person's genome in order to find or verify ancestral genealogical relationships, or to estimate the ethnic mixture of an individual. Since different testing companies use different ethnic reference groups and different matching algorithms, ethnicity estimates for an individual vary between tests, sometimes dramatically.
A Y-STR is a short tandem repeat (STR) on the Y-chromosome. Y-STRs are often used in forensics, paternity, and genealogical DNA testing. Y-STRs are taken specifically from the male Y chromosome. These Y-STRs provide a weaker analysis than autosomal STRs because the Y chromosome is only found in males, which are only passed down by the father, making the Y chromosome in any paternal line practically identical. This causes a significantly smaller amount of distinction between Y-STR samples. Autosomal STRs provide a much stronger analytical power because of the random matching that occurs between pairs of chromosomes during the zygote-making process.

Haplogroup G (M201) is a human Y-chromosome haplogroup. It is one of two branches of the parent haplogroup GHIJK, the other being HIJK.

In human genetics, a human Y-chromosome DNA haplogroup is a haplogroup defined by mutations in the non-recombining portions of DNA from the male-specific Y chromosome. Many people within a haplogroup share similar numbers of short tandem repeats (STRs) and types of mutations called single-nucleotide polymorphisms (SNPs).
In genetic genealogy, a unique-event polymorphism (UEP) is a genetic marker that corresponds to a mutation that is likely to occur so infrequently that it is believed overwhelmingly probable that all the individuals who share the marker, worldwide, will have inherited it from the same common ancestor, and the same single mutation event.
Haplogroup G2b-M377 is a Y-chromosome haplogroup and is defined by the presence of the M377 mutation. It is a branch of Haplogroup G2, which in turn is defined by the presence of the M201 mutation.
The Sorenson Molecular Genealogy Foundation (SMGF) was an independent DNA and genealogical research institution with the goal of demonstrating how the peoples of the world are related. SMGF collected DNA samples and genealogical information from individuals across the globe to establish these connections.

In human genetics, Haplogroup G-M285 or G-M342, also known as Haplogroup G1, is a Y-chromosome haplogroup. Haplogroup G1 is a primary subclade of haplogroup G.
Haplogroup G-FGC7535, also known as Haplogroup G2a1, is a Y-chromosome haplogroup. It is an immediate descendant of G2a (G-P15), which is a primary branch of haplogroup G2 (P287).
In human genetics, Haplogroup G-P303 is a Y-chromosome haplogroup. It is a branch of haplogroup G (Y-DNA) (M201). In descending order, G-P303 is additionally a branch of G2 (P287), G2a (P15), G2a2, G2a2b, G2a2b2, and finally G2a2b2a. This haplogroup represents the majority of haplogroup G men in most areas of Europe west of Russia and the Black Sea. To the east, G-P303 is found among G persons across the Middle East, Iran, the southern Caucasus area, China, and India. G-P303 exhibits its highest diversity in the Levant.
The relationship of the Mayas to other indigenous peoples of the Americas has been assessed using traditional genetic markers. Mayas inhabited several parts of Mexico and Central America, including Chiapas, the northern lowlands of the Yucatán Peninsula, the southern lowlands and highlands of Guatemala, Belize, and parts of western El Salvador and Honduras. Genetic studies of the Maya people are reported to show higher levels of variation when compared to other groups.



