Related Research Articles

Bioinformatics is an interdisciplinary field of science that develops methods and software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, chemistry, physics, computer science, computer programming, information engineering, mathematics and statistics to analyze and interpret biological data. The subsequent process of analyzing and interpreting data is referred to as computational biology.

The proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. Proteomics is the study of the proteome.

Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body.
In bioinformatics, sequence analysis is the process of subjecting a DNA, RNA or peptide sequence to any of a wide range of analytical methods to understand its features, function, structure, or evolution. It can be performed on the entire genome, transcriptome or proteome of an organism, and can also involve only selected segments or regions, like tandem repeats and transposable elements. Methodologies used include sequence alignment, searches against biological databases, and others.
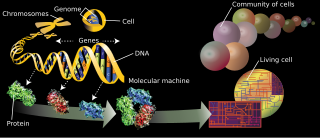
The branches of science known informally as omics are various disciplines in biology whose names end in the suffix -omics, such as genomics, proteomics, metabolomics, metagenomics, phenomics and transcriptomics. Omics aims at the collective characterization and quantification of pools of biological molecules that translate into the structure, function, and dynamics of an organism or organisms.

Functional genomics is a field of molecular biology that attempts to describe gene functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects. Functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "candidate-gene" approach.
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules but can also describe sets of indirect interactions among genes.
The completion of the human genome sequencing in the early 2000s was a turning point in genomics research. Scientists have conducted series of research into the activities of genes and the genome as a whole. The human genome contains around 3 billion base pairs nucleotide, and the huge quantity of data created necessitates the development of an accessible tool to explore and interpret this information in order to investigate the genetic basis of disease, evolution, and biological processes. The field of genomics has continued to grow, with new sequencing technologies and computational tool making it easier to study the genome.

The Generic Model Organism Database (GMOD) project provides biological research communities with a toolkit of open-source software components for visualizing, annotating, managing, and storing biological data. The GMOD project is funded by the United States National Institutes of Health, National Science Foundation and the USDA Agricultural Research Service.

Microarray analysis techniques are used in interpreting the data generated from experiments on DNA, RNA, and protein microarrays, which allow researchers to investigate the expression state of a large number of genes – in many cases, an organism's entire genome – in a single experiment. Such experiments can generate very large amounts of data, allowing researchers to assess the overall state of a cell or organism. Data in such large quantities is difficult – if not impossible – to analyze without the help of computer programs.

Galaxy is a scientific workflow, data integration, and data and analysis persistence and publishing platform that aims to make computational biology accessible to research scientists that do not have computer programming or systems administration experience. Although it was initially developed for genomics research, it is largely domain agnostic and is now used as a general bioinformatics workflow management system.

MicrobesOnline is a publicly and freely accessible website that hosts multiple comparative genomic tools for comparing microbial species at the genomic, transcriptomic and functional levels. MicrobesOnline was developed by the Virtual Institute for Microbial Stress and Survival, which is based at the Lawrence Berkeley National Laboratory in Berkeley, California. The site was launched in 2005, with regular updates until 2011.
BIOBASE is an international bioinformatics company headquartered in Wolfenbüttel, Germany. The company focuses on the generation, maintenance, and licensing of databases in the field of molecular biology, and their related software platforms.
GeneCards is a database of human genes that provides genomic, proteomic, transcriptomic, genetic and functional information on all known and predicted human genes. It is being developed and maintained by the Crown Human Genome Center at the Weizmann Institute of Science, in collaboration with LifeMap Sciences.

Proteogenomics is a field of biological research that utilizes a combination of proteomics, genomics, and transcriptomics to aid in the discovery and identification of peptides. Proteogenomics is used to identify new peptides by comparing MS/MS spectra against a protein database that has been derived from genomic and transcriptomic information. Proteogenomics often refers to studies that use proteomic information, often derived from mass spectrometry, to improve gene annotations. The utilization of both proteomics and genomics data alongside advances in the availability and power of spectrographic and chromatographic technology led to the emergence of proteogenomics as its own field in 2004.
The Multi-Omics Profiling Expression Database (MOPED) was an expanding multi-omics resource that supports rapid browsing of transcriptomics and proteomics information from publicly available studies on model organisms and humans. As to date (2021) it has ceased activities and is unaccessible online.

Gene set enrichment analysis (GSEA) (also called functional enrichment analysis or pathway enrichment analysis) is a method to identify classes of genes or proteins that are over-represented in a large set of genes or proteins, and may have an association with different phenotypes (e.g. different organism growth patterns or diseases). The method uses statistical approaches to identify significantly enriched or depleted groups of genes. Transcriptomics technologies and proteomics results often identify thousands of genes, which are used for the analysis.
The Expression Atlas is a database maintained by the European Bioinformatics Institute that provides information on gene expression patterns from RNA-Seq and Microarray studies, and protein expression from Proteomics studies. The Expression Atlas allows searches by gene, splice variant, protein attribute, disease, treatment or organism part. Individual genes or gene sets can be searched for. All datasets in Expression Atlas have its metadata manually curated and its data analysed through standardised analysis pipelines. There are two components to the Expression Atlas, the Baseline Atlas and the Differential Atlas:

The Centre for Genomic Regulation is a biomedical and genomics research centre based on Barcelona. Most of its facilities and laboratories are located in the Barcelona Biomedical Research Park, in front of Somorrostro beach.
References
- ↑ Kolker E, Higdon R, Morgan P, Sedensky M, Welch D, Bauman A, Stewart E, Haynes W, Broomall W, Kolker N (December 2011). "SPIRE: Systematic protein investigative research environment". J Proteomics. 75 (1): 122–6. doi:10.1016/j.jprot.2011.05.009. PMID 21609792.