
C11orf49 is a protein coding gene that in humans encodes for the C11orf49 protein. It is heavily expressed in brain tissue and peripheral blood mononuclear cells, with the latter being an important component of the immune system. It is predicted that the C11orf49 protein acts as a kinase, and has been shown to interact with HTT and APOE2.

Family with sequence similarity 63, member A is a protein that, is encoded by the FAM63A gene in humans,. It is located on the minus strand of chromosome 1 at locus 1q21.3.
C5orf34 is a protein that in humans is encoded by the C5orf34 gene (5p12).
WD repeat-containing protein 90 is a protein that, in humans, is encoded by the WDR90 gene (16p13.3). This human protein is 1750 amino acids, and has a molecular weight of 187.7 kDa. It contains multiple WD40 repeat domains and one domain of unknown function. This protein is conserved all the way back to invertebrates. Proteins containing WD transducin repeating domains have been found to play a role in a variety of functions ranging from signal transduction and transcription regulation to cell cycle control, autophagy and apoptosis.

PRR29 is a protein encoded by the PRR29 gene located in humans on chromosome 17 at 17q23.

Transmembrane Protein 176B, or TMEM176B is a transmembrane protein that in humans is encoded by the TMEM176B gene. It is thought to play a role in the process of maturation of dendritic cells.
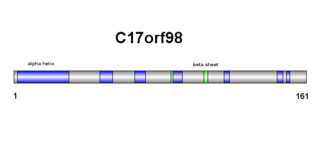
C17orf98 is a protein which in humans is coded by the gene c17orf98. The protein is derived from Homo sapiens chromosome 17. The C17orf98 gene consists of a 6,302 base sequence. Its mRNA has three exons and no alternative splice sites. The protein has 154 amino acids, with no abnormal amino acid levels. C17orf98 has a domain of unknown function (DUF4542) and is 17.6kDa in weight. C17orf98 does not belong to any other families nor does it have any isoforms. The protein has orthologs with high percent similarity in mammals and reptiles. The protein has additional distantly related orthologs across the metazoan kingdom, culminating with the sponge family.

TMEM44 is a protein that in humans is encoded by the TMEM44 gene. DKFZp686O18124 is a synonym of TMEM44.

Chromosome 4 open reading frame 51 (C4orf51) is a protein which in humans is encoded by the C4orf51 gene.

Cilia- and flagella-associated protein 299 (CFAP299), is a protein that in humans is encoded by the CFAP299 gene. CFAP299 is predicted to play a role in spermatogenesis and cell apoptosis.

Single-pass membrane and coiled-coil domain-containing protein 3 is a protein that is encoded in humans by the SMCO3 gene.

C5orf46 is a protein coding gene located on chromosome 5 in humans. It is also known as sssp1, or skin and saliva secreted protein 1. There are two known isoforms known in humans, with isoform 2 being the longer of the two. The protein encoded is predicted to have one transmembrane domain, and has a predicted molecular weight of 9,692 Da, and a basal isoelectric point of 4.67.

C16orf90 or chromosome 16 open reading frame 90 produces uncharacterized protein C16orf90 in homo sapiens. C16orf90's protein has four predicted alpha-helix domains and is mildly expressed in the testes and lowly expressed throughout the body. While the function of C16orf90 is not yet well understood by the scientific community, it has suspected involvement in the biological stress response and apoptosis based on expression data from microarrays and post-translational modification data.

Transmembrane protein 221 (TMEM221) is a protein that in humans is encoded by the TMEM221 gene. The function of TMEM221 is currently not well understood.
TMEM275 is a protein that in humans is encoded by the TMEM275 gene. TMEM275 has two, highly-conserved, helical trans-membrane regions. It is predicted to reside within the plasma membrane or the endoplasmic reticulum's membrane.
C3orf56 is a protein encoding gene found on chromosome 3. Although, the structure and function of the protein is not well understood, it is known that the C3orf56 protein is exclusively expressed in metaphase II of oocytes and degrades as the oocyte develops towards the blastocyst stage. Degradation of the C3orf56 protein suggests that this gene plays a role in the progression from maternal to embryonic genome and in embryonic genome activation.

Chromosome 9 open reading frame 85, commonly known as C9orf85, is a protein in Homo sapiens encoded by the C9orf85 gene. The gene is located at 9q21.13. When spliced, four different isoforms are formed. C9orf85 has a predicted molecular weight of 20.17 kdal. Isoelectric point was found to be 9.54. The function of the gene has not yet been confirmed, however it has been found to show high levels of expression in cells of high differentiation.

The FAM214B, also known as protein family with sequence similarity 214, B (FAM214B) is a protein that, in humans, is encoded by the FAM214B gene located on the human chromosome 9. The protein has 538 amino acids. The gene contain 9 exon. There has been studies that there are low expression of this gene in patients with major depression disorder. In most organisms such as mammals, amphibians, reptiles, and birds, there are high levels of gene expression in the bone marrow and blood. For humans in fetal development, FAM214B is mostly expressed in the brains and bone marrow.

PANO1 is a protein which in humans is encoded by the PANO1 gene. PANO1 is an apoptosis inducing protein that is able to regulate the function of tumor suppressor. More specifically, P14ARF is a protein in which in humans is modulated by the PANO1 gene. P14ARF is known to function as a tumor suppressor. When PANO1 is highly expressed in the cells, it is able to modulate p14ARF by stabilizing it and protecting it from degradation. With a confidence level of 5 out of 5, PANO1 has been theorized to be expressed in the nucleolus of the cell. PANO1 is an intron-less gene. Intron-less genes only make up about 3% of the human genome. A functional analysis of these types of genes revealed that they often have tissue-specific expression in tissues such as the nervous system and testis. This kind of expression is commonly associated with neuropathies, disease, and cancer. The tissue types that PANO1 has the highest expression in, are the cerebellum regions of the brain as well as pituitary and testis tissues.

C11orf98 is a protein-encoding gene on chromosome 11 in humans of unknown function. It is otherwise known as c11orf48. The gene spans the chromosomal locus from 62,662,817-62,665,210. There are 4 exons. It spans across 2,394 base pairs of DNA and produces an mRNA that is 646 base pairs long.















