
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA . Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism.

In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication.
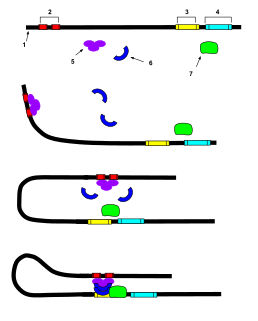
In genetics, an enhancer is a short region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are cis-acting. They can be located up to 1 Mbp away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes.
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses.
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology.
In molecular biology, the TATA box is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence.

Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.
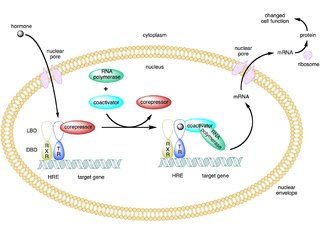
A coactivator is a type of transcriptional coregulator that binds to an activator to increase the rate of transcription of a gene or set of genes. The activator contains a DNA binding domain that binds either to a DNA promoter site or a specific DNA regulatory sequence called an enhancer. Binding of the activator-coactivator complex increases the speed of transcription by recruiting general transcription machinery to the promoter, therefore increasing gene expression. The use of activators and coactivators allows for highly specific expression of certain genes depending on cell type and developmental stage.
Cis-regulatory elements (CREs) or Cis-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology.

Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.
In molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that repress are known as corepressors. The mechanism of action of transcription coregulators is to modify chromatin structure and thereby make the associated DNA more or less accessible to transcription. In humans several dozen to several hundred coregulators are known, depending on the level of confidence with which the characterisation of a protein as a coregulator can be made. One class of transcription coregulators modifies chromatin structure through covalent modification of histones. A second ATP dependent class modifies the conformation of chromatin.

Transcriptional repressor CTCF also known as 11-zinc finger protein or CCCTC-binding factor is a transcription factor that in humans is encoded by the CTCF gene. CTCF is involved in many cellular processes, including transcriptional regulation, insulator activity, V(D)J recombination and regulation of chromatin architecture.

In the field of molecular biology, a corepressor is a molecule that represses the expression of genes. In prokaryotes, corepressors are small molecules whereas in eukaryotes, corepressors are proteins. A corepressor does not directly bind to DNA, but instead indirectly regulates gene expression by binding to repressors.

Myocyte-specific enhancer factor 2A is a protein that in humans is encoded by the MEF2A gene. MEF2A is a transcription factor in the Mef2 family. In humans it is located on chromosome 15q26. Certain mutations in MEF2A cause an autosomal dominant form of coronary artery disease and myocardial infarction.

Nuclear transcription factor Y subunit gamma is a protein that in humans is encoded by the NFYC gene.

Carbohydrate-responsive element-binding protein (ChREBP) also known as MLX-interacting protein-like (MLXIPL) is a protein that in humans is encoded by the MLXIPL gene. The protein name derives from the protein's interaction with carbohydrate response element sequences of DNA.
Chromatin Interaction Analysis by Paired-End Tag Sequencing is a technique that incorporates chromatin immunoprecipitation (ChIP)-based enrichment, chromatin proximity ligation, Paired-End Tags, and High-throughput sequencing to determine de novo long-range chromatin interactions genome-wide.
RegulonDB is a database of the regulatory network of gene expression in Escherichia coli K-12. RegulonDB also models the organization of the genes in transcription units, operons and regulons. A total of 120 sRNAs with 231 total interactions which all together regulate 192 genes are also included. RegulonDB was founded in 1998 and also contributes data to the EcoCyc database.
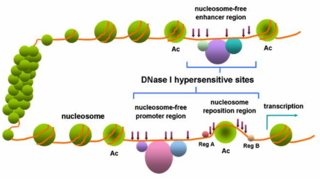
In genetics, DNase I hypersensitive sites (DHSs) are regions of chromatin that are sensitive to cleavage by the DNase I enzyme. In these specific regions of the genome, chromatin has lost its condensed structure, exposing the DNA and making it accessible. This raises the availability of DNA to degradation by enzymes, such as DNase I. These accessible chromatin zones are functionally related to transcriptional activity, since this remodeled state is necessary for the binding of proteins such as transcription factors.












