
The Acido-1 RNA motif is a conserved RNA structure identified by bioinformatics. It is found only in acidobacteriota, and appears to be a non-coding RNA as it does not have a consistent association with protein-coding genes.
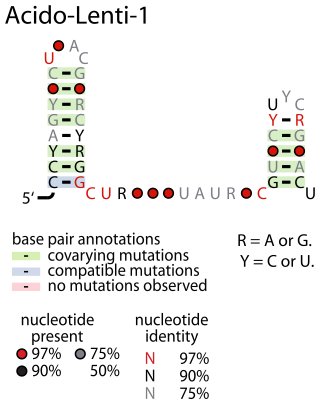
The Acido-Lenti-1 RNA motif describes a predicted non-coding RNA that is found in bacteria within the phyla acidobacteriota and lentisphaerota. It is sometimes found nearby to group II introns, but the reason for this apparent association is unknown.

The Bacteroides-1 RNA motif is a conserved RNA structure identified in bacteria within the genus Bacteroides. The RNAs are typically found downstream of of genes that participate in the synthesis of exopolysaccharides of unknown types. It is possible that Bacteroides-1 RNAs regulate the upstream genes, but since this mode of regulation is unusual in bacteria, it is more likely that the structure functions as a non-coding RNA.
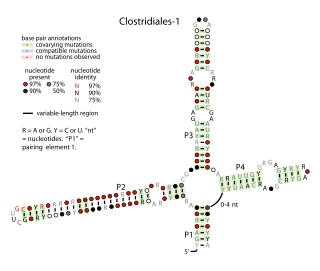
The Clostridiales-1 RNA motif is a conserved RNA structure identified by bioinformatics. It is a four-stem structure common in bacteria that inhabit the human gut and is also found in a variety of bacteria classified within the order Clostridiales. Its function is unknown.
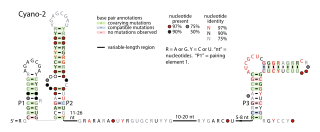
The Cyano-2 RNA motif is a conserved RNA structure identified by bioinformatics. Cyano-2 RNAs are found in Cyanobacterial species classified within the genus Synechococcus. Many terminal loops in the two conserved stem-loops contain the nucleotide sequence GCGA, and these sequences might in some cases form stable GNRA tetraloops. Since the two stem-loops are somewhat distant from one another it is possible that they represent two independent non-coding RNAs that are often or always co-transcribed. The region one thousand base pairs upstream of predicted Cyano-2 RNAs is usually devoid of annotated features such as RNA or protein-coding genes. This absence of annotated genes within one thousand base pairs is relatively unusual within bacteria.

The Gut-1 RNA motif is a conserved RNA structure identified by bioinformatics. These RNAs are present in environmental sequences, and as of 2010 are not known to be present in any species that has been grown under laboratory conditions. Gut-1 RNA is exclusively found in DNA from uncultivated bacteria present in samples from the human gut.

The L17 downstream element RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. All known L17 downstream elements were detected immediately downstream of genes encoding the L17 subunit of the ribosome, and therefore might be in the 3' untranslated regions of these genes. The element is found in a variety of lactic acid bacteria and in the genus Listeria.
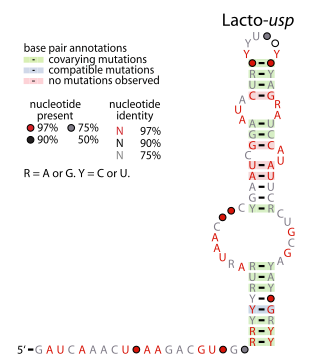
The Lacto-usp RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. Lacto-usp RNAs are found exclusively in lactic acid bacteria, and exclusively in the possible 5′ untranslated regions of operons that contain a hypothetical gene and a usp gene. The usp gene encodes the universal stress protein. It was proposed that the Lacto-usp might correspond to the 6S RNA of the relevant species, because four of five of these species lack a predicted 6S RNA, and 6S RNAs commonly occur in 5′ UTRs of usp genes. However, given that the Lacto-usp RNA motif is much shorter than the standard 6S RNA structure, the function of Lacto-usp RNAs remains unclear.

The leu/phe-leader RNA motif is a conserved RNA structure identified by bioinformatics. These RNAs function as peptide leaders. They contain a short open reading frame (ORF) that contains many codons for leucine or phenylalanine. Normally, expression of the downstream genes is suppressed. However, when cellular concentrations of the relevant amino acid is low, ribosome stalling leads to an alternate structure that enables downstream gene expression.
The Lnt RNA motif refers to a conserved RNA structure found in certain bacteria. Specifically, Lnt RNAs are known only in species within the phylum Chlorobiota, and are located in the possible 5' untranslated regions of genes that are annotated as encoding apolipoprotein N-acyltransferase enzymes. There is some doubt as to whether the indicated motif is transcribed as RNA, or whether its reverse complement is transcribed. If the reverse complement is transcribed it would potentially in 5' UTRs of genes encoding bacteriochlorophyll A, and would be close to the start codon of those genes.

The psbNH RNA motif describes a class of RNA molecules that have a conserved secondary structure. psbNH RNAs are always found between psbH and psbH genes, both of which are involved in the cyanobacterial photosystem II are transcribed in opposite orientations. It is unknown whether the biological psbNH RNA is as depicted in the diagram, or whether its reverse complement is the transcribed molecule. In either case, the RNA would be in the 5' untranslated region of a gene, either psbN or psbH, and likely a cis-regulatory element.

The Pseudomon-Rho RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. The RNAs that conform to this motif are found in species within the genus Pseudomonas, as well as the related Azotobacter vinelandii. They are consistently located in what could be the 5' untranslated regions of genes that encode the Rho factor protein, and this arrangement in bacteria suggested that Pseudomon-Rho RNAs might be cis-regulatory elements that regulate concentrations of the Rho protein.

The Rhizobiales-2 RNA motif is a set of RNAs found in certain bacteria that are presumed to be homologous because they conserve a common primary and secondary structure. The motif was discovered using bioinformatics, and is found only within bacteria that belong to the order Hyphomicrobiales, in turn a kind of alphaproteobacteria. Because Rhizobiales-2 RNAs are not consistently located in proximity to genes of a consistent class or function, these RNAs are presumed to function as non-coding RNAs.

The SAM-Chlorobi RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNAs are found only in bacteria classified as within the phylum Chlorobiota. These RNAs are always in the 5' untranslated regions of operons that contain metK and ahcY genes. metK genes encode methionine adenosyltransferase, which synthesizes S-adenosyl methionine (SAM), and ahcY genes encode S-adenosylhomocysteine hydrolase, which degrade the related metabolite S-Adenosyl-L-homocysteine (SAH). In fact all predicted metK and ahcY genes within Chlorobiota bacteria as of 2010 are preceded by predicted SAM-Chlorobi RNAs. Predicted promoter sequences are consistently found upstream of SAM-Chlorobi RNAs, and these promoter sequences imply that SAM-Chlorobi RNAs are indeed transcribed as RNAs. The promoter sequences are commonly associated with strong transcription in the phyla Chlorobiota and Bacteroidota, but are not used by most lineages of bacteria. The placement of SAM-Chlorobi RNAs suggests that they are involved in the regulation of the metK/ahcY operon through an unknown mechanism.
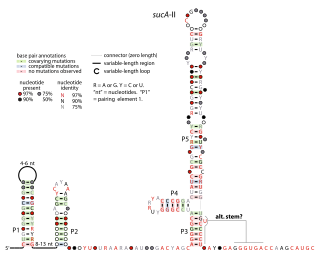
The sucA-II RNA motif is a conserved RNA structure identified by bioinformatics. It is consistently found in the presumed 5' untranslated regions of sucA genes, which encode Oxoglutarate dehydrogenase enzymes that participate in the citric acid cycle. Given this arrangement, sucA-II RNAs might regulate the downstream sucA gene. This genetic arrangement is similar to the previously reported sucA RNA motif. However, sucA-II RNAs are found only in bacteria classified within the genus Pseudomonas, whereas the previously reported motif is found only in betaproteobacteria.

The Termite-flg RNA motif is a conserved RNA structure identified by bioinformatics. Genomic sequences corresponding to Termite-flg RNAs have been identified only in uncultivated bacteria present in the termite hindgut. As of 2010 it has not been identified in the DNA of any cultivated species, and is thus an example of RNAs present in environmental samples.
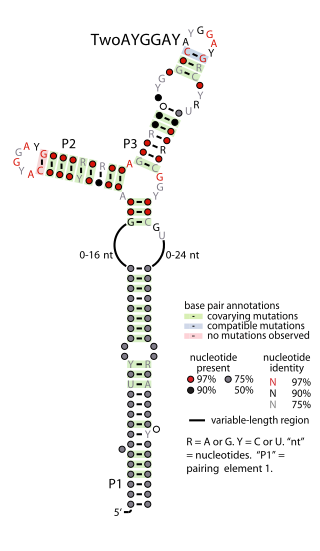
The TwoAYGGAY RNA motif is a conserved RNA structure identified by bioinformatics. Its name refers to the conserved AYGGAY nucleotide sequence found in the motif's two terminal loops. The RNAs are found in sequences derived from DNA extracted from uncultivated bacteria present in the human gut, as well as some bacteria in the classes Clostridia and Gammaproteobacteria.

The Whalefall-1 RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. Structurally, the motif consists of two stem-loops, the second of which is often terminated by a CUUG tetraloop, which is an energetically favorable RNA sequence. Whalefall-1 RNAs are found only in DNA extracted from uncultivated bacteria found on whale fall, i.e., a whale carcass. As of 2010, Whalefall-1 RNAs have not been detected in any known, cultivated species of bacteria, and are thus one of several RNAs present in environmental samples.
The Ocean-V RNA motif is a conserved RNA structure discovered using bioinformatics. Only a few Ocean-V RNA sequences have been detected, all in sequences derived from DNA that was extracted from uncultivated bacteria found in ocean water. As of 2010, no Ocean-V RNA has been detected in any known, cultivated organism.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Bacillota and Gammaproteobacteria.


















