
Bleomycin is a medication used to treat cancer. This includes Hodgkin's lymphoma, non-Hodgkin's lymphoma, testicular cancer, ovarian cancer, and cervical cancer among others. Typically used with other cancer medications, it can be given intravenously, by injection into a muscle or under the skin. It may also be administered inside the chest to help prevent the recurrence of a fluid around the lung due to cancer; however talc is better for this.
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be made by bacteria inside these organisms. While there exist a wide range of peptides that are not synthesized by ribosomes, the term nonribosomal peptide typically refers to a very specific set of these as discussed in this article.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.
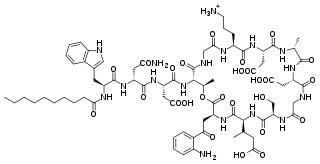
Daptomycin, sold under the brand name Cubicin among others, is a lipopeptide antibiotic used in the treatment of systemic and life-threatening infections caused by Gram-positive organisms.
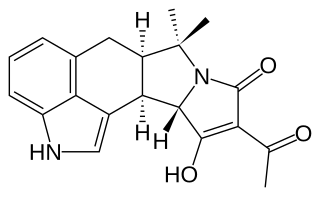
Cyclopiazonic acid (α-CPA), a mycotoxin and a fungal neurotoxin, is made by the molds Aspergillus and Penicillium. It is an indole-tetramic acid that serves as a toxin due to its ability to inhibit calcium-dependent ATPases found in the endoplasmic and sarcoplasmic reticulum. This inhibition disrupts the muscle contraction-relaxation cycle and the calcium gradient that is maintained for proper cellular activity in cells.
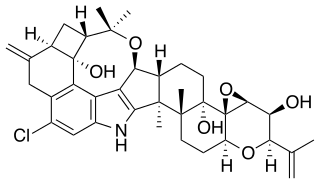
Penitrem A (tremortin) is an indole-diterpenoid mycotoxin produced by certain species of Aspergillus, Claviceps, and Penicillium, which can be found growing on various plant species such as ryegrass. Penitrem A is one of many secondary metabolites following the synthesis of paxilline in Penicillium crostosum. Penitrem A poisoning in humans and animals usually occurs through the consumption of contaminated foods by mycotoxin-producing species, which is then distributed through the body by the bloodstream. It bypasses the blood-brain barrier to exert its toxicological effects on the central nervous system. In humans, penitrem A poisoning has been associated with severe tremors, hyperthermia, nausea/vomiting, diplopia, and bloody diarrhea. In animals, symptoms of penitrem A poisoning has been associated with symptoms ranging from tremors, seizures, and hyperthermia to ataxia and nystagmus.
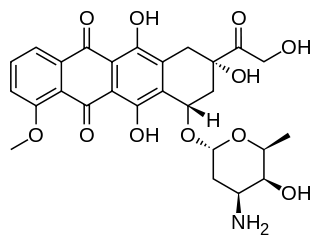
Doxorubicin (DXR) is a 14-hydroxylated version of daunorubicin, the immediate precursor of DXR in its biosynthetic pathway. Daunorubicin is more abundantly found as a natural product because it is produced by a number of different wild type strains of streptomyces. In contrast, only one known non-wild type species, streptomyces peucetius subspecies caesius ATCC 27952, was initially found to be capable of producing the more widely used doxorubicin. This strain was created by Arcamone et al. in 1969 by mutating a strain producing daunorubicin, but not DXR, at least in detectable quantities. Subsequently, Hutchinson's group showed that under special environmental conditions, or by the introduction of genetic modifications, other strains of streptomyces can produce doxorubicin. His group has also cloned many of the genes required for DXR production, although not all of them have been fully characterized. In 1996, Strohl's group discovered, isolated and characterized dox A, the gene encoding the enzyme that converts daunorubicin into DXR. By 1999, they produced recombinant Dox A, a Cytochrome P450 oxidase, and found that it catalyzes multiple steps in DXR biosynthesis, including steps leading to daunorubicin. This was significant because it became clear that all daunorubicin producing strains have the necessary genes to produce DXR, the much more therapeutically important of the two. Hutchinson's group went on to develop methods to improve the yield of DXR, from the fermentation process used in its commercial production, not only by introducing Dox A encoding plasmids, but also by introducing mutations to deactivate enzymes that shunt DXR precursors to less useful products, for example baumycin-like glycosides. Some triple mutants, that also over-expressed Dox A, were able to double the yield of DXR. This is of more than academic interest because at that time DXR cost about $1.37 million per kg and current production in 1999 was 225 kg per annum. More efficient production techniques have brought the price down to $1.1 million per kg for the non-liposomal formulation. Although DXR can be produced semi-synthetically from daunorubicin, the process involves electrophilic bromination and multiple steps and the yield is poor. Since daunorubicin is produced by fermentation, it would be ideal if the bacteria could complete DXR synthesis more effectively.

Ergocryptine is an ergopeptine and one of the ergot alkaloids. It is isolated from ergot or fermentation broth and it serves as starting material for the production of bromocriptine.
Streptogramin A is a group of antibiotics within the larger family of antibiotics known as streptogramins. They are synthesized by the bacteria Streptomyces virginiae. The streptogramin family of antibiotics consists of two distinct groups: group A antibiotics contain a 23-membered unsaturated ring with lactone and peptide bonds while group B antibiotics are depsipeptides. While structurally different, these two groups of antibiotics act synergistically, providing greater antibiotic activity than the combined activity of the separate components. These antibiotics have until recently been commercially manufactured as feed additives in agriculture, although today there is increased interest in their ability to combat antibiotic-resistant bacteria, particularly vancomycin-resistant bacteria.
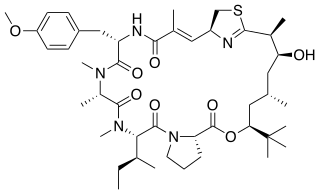
Apratoxin A - is a cyanobacterial secondary metabolite, known as a potent cytotoxic marine natural product. It is a derivative of the Apratoxin family of cytotoxins. The mixed peptide-polyketide natural product comes from a polyketide synthase/non-ribosomal peptide synthase pathway (PKS/NRPS). This cytotoxin is known for inducing G1-phase cell cycle arrest and apoptosis. This natural product's activity has made it a popular target for developing anticancer derivatives.
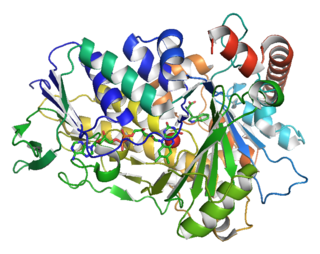
Tryptophan 7-halogenase (EC 1.14.19.9, PrnA, RebH) is an enzyme with systematic name L-tryptophan:FADH2 oxidoreductase (7-halogenating). This enzyme catalyses the following chemical reaction:

Hectochlorin is a lipopeptide that exhibits potent antifungal activity against C. albicans and a number of plants pathogens, as well as inhibiting growth of human cell lines by hyperpolymerization of actin. It was originally isolated from the filamentous cyanobacterium Moorea producens JHB, collected from Hector Bay, Jamaica, 1996, which is a strain also known for being the producer of other two potent biomolecules named Jamaicamide A and Cryptomaldamide. Due to its activity against plants pathogens, synthetic efforts elucidated the compound’s total synthesis in 2002. Moorea species are normally the main component of the dietary of some sea hares, which concentrate the cyanobacterial metabolites as a mechanism of defense from predators. Therefore, in 2005, hectochlorin was re-isolated from the Thai sea hare Bursatella leachii, along with a new analogue, deacetylhectochlorin. Another reisolation of hectochlorin was reported in 2013, from another Moorea producens strain (RS05), isolated from the Red Sea, surprising in a non-tropical environment, as opposed to the other Moorea strains isolated before. The predicted biosynthesis of hectochlorin was published in 2007 and consists in a hybrid NRPS-PKS, with a hexanoic acid as start unit that becomes halogenated twice in the position 5, producing fairly rare gem-dichloro group, that along with two 2,3-dihydroxyisovaleric acid (DHIV) units compose a very interesting bioactive molecule.

Pseurotin A is a secondary metabolite of Aspergillus.

Bottromycin is a macrocyclic peptide with antibiotic activity. It was first discovered in 1957 as a natural product isolated from Streptomyces bottropensis. It has been shown to inhibit methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci (VRE) among other Gram-positive bacteria and mycoplasma. Bottromycin is structurally distinct from both vancomycin, a glycopeptide antibiotic, and methicillin, a beta-lactam antibiotic.
Spectinabilin is a nitrophenyl-substituted polyketide metabolite. It was first isolated from crude streptovaricin complex produced by Streptomyces spectabilis and presented at the 13th Interscience Conference on Antimicrobial Agents and Chemotherapy in Washington, D.C. in September 1973. Spectinabilin is a biologically active compound, exhibiting both antimalarial and antiviral activity.

The flavin-containing monooxygenase (FMO) protein family specializes in the oxidation of xeno-substrates in order to facilitate the excretion of these compounds from living organisms. These enzymes can oxidize a wide array of heteroatoms, particularly soft nucleophiles, such as amines, sulfides, and phosphites. This reaction requires an oxygen, an NADPH cofactor, and an FAD prosthetic group. FMOs share several structural features, such as a NADPH binding domain, FAD binding domain, and a conserved arginine residue present in the active site. Recently, FMO enzymes have received a great deal of attention from the pharmaceutical industry both as a drug target for various diseases and as a means to metabolize pro-drug compounds into active pharmaceuticals. These monooxygenases are often misclassified because they share activity profiles similar to those of cytochrome P450 (CYP450), which is the major contributor to oxidative xenobiotic metabolism. However, a key difference between the two enzymes lies in how they proceed to oxidize their respective substrates; CYP enzymes make use of an oxygenated heme prosthetic group, while the FMO family utilizes FAD to oxidize its substrates.
Fostriecin is a type I polyketide synthase (PKS) derived natural product, originally isolated from the soil bacterium Streptomyces pulveraceus. It belongs to a class of natural products which characteristically contain a phosphate ester, an α,β-unsaturated lactam and a conjugated linear diene or triene chain produced by Streptomyces. This class includes structurally related compounds cytostatin and phoslactomycin. Fostriecin is a known potent and selective inhibitor of protein serine/threonine phosphatases, as well as DNA topoisomerase II. Due to its activity against protein phosphatases PP2A and PP4 which play a vital role in cell growth, cell division, and signal transduction, fostriecin was looked into for its antitumor activity in vivo and showed in vitro activity against leukemia, lung cancer, breast cancer, and ovarian cancer. This activity is thought to be due to PP2A's assumed role in regulating apoptosis of cells by activating cytotoxic T-lymphocytes and natural killer cells involved in tumor surveillance, along with human immunodeficiency virus-1 (HIV-1) transcription and replication.

Annimycin (4-(Z)-annimycin) is a polyenoic acid amide natural product produced by Streptomyces calvus. Annimycin inhibits the sporulation of several actinobacterial genera.

Dihydromaltophilin, or heat stable anti-fungal factor (HSAF), is a secondary metabolite of Streptomyces sp. and Lysobacter enzymogenes. HSAF is a polycyclic tetramate lactam containing a single tetramic acid unit and a 5,5,6-tricyclic system. HSAF has been shown to have anti-fungal activity mediated through the disruption of the biosynthesis of Sphingolipid's by targeting a ceramide synthase unique to fungi.
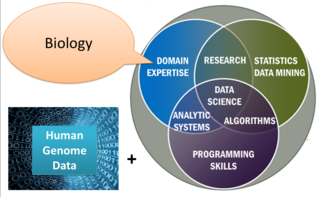
Genome mining describes the exploitation of genomic information for the discovery of biosynthetic pathways of natural products and their possible interactions. It depends on computational technology and bioinformatics tools. The mining process relies on a huge amount of data accessible in genomic databases. By applying data mining algorithms, the data can be used to generate new knowledge in several areas of medicinal chemistry, such as discovering novel natural products.



















