
Flavivirus is a genus of positive-strand RNA viruses in the family Flaviviridae. The genus includes the West Nile virus, dengue virus, tick-borne encephalitis virus, yellow fever virus, Zika virus and several other viruses which may cause encephalitis, as well as insect-specific flaviviruses (ISFs) such as cell fusing agent virus (CFAV), Palm Creek virus (PCV), and Parramatta River virus (PaRV). While dual-host flaviviruses can infect vertebrates as well as arthropods, insect-specific flaviviruses are restricted to their competent arthropods. The means by which flaviviruses establish persistent infection in their competent vectors and cause disease in humans depends upon several virus-host interactions, including the intricate interplay between flavivirus-encoded immune antagonists and the host antiviral innate immune effector molecules.

Defective interfering particles (DIPs), also known as defective interfering viruses, are spontaneously generated virus mutants in which a critical portion of the particle's genome has been lost due to defective replication or non-homologous recombination. The mechanism of their formation is presumed to be as a result of template-switching during replication of the viral genome, although non-replicative mechanisms involving direct ligation of genomic RNA fragments have also been proposed. DIPs are derived from and associated with their parent virus, and particles are classed as DIPs if they are rendered non-infectious due to at least one essential gene of the virus being lost or severely damaged as a result of the defection. A DIP can usually still penetrate host cells, but requires another fully functional virus particle to co-infect a cell with it, in order to provide the lost factors.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.

Tombusviridae is a family of single-stranded positive sense RNA plant viruses. There are three subfamilies, 17 genera, and 95 species in this family. The name is derived from Tomato bushy stunt virus (TBSV).
Tombusvirus is a genus of viruses, in the family Tombusviridae. Plants serve as natural hosts. There are 17 species in this genus. Symptoms associated with this genus include mosaic. The name of the genus comes from Tomato bushy stunt virus.
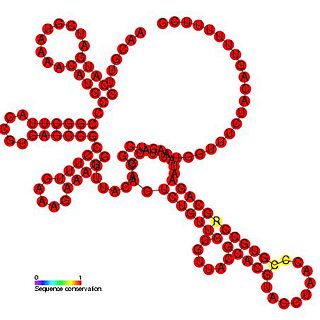
The Bamboo mosaic potexvirus (BaMV) cis-regulatory element represents a cloverleaf-like cis-regulatory element found in the 3' UTR of the bamboo mosaic virus. This family is thought to play an important role in the initiation of minus-strand RNA synthesis and may also be involved in the regulation of viral replication.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.

The Infectious bronchitis virus D-RNA is an RNA element known as defective RNA or D-RNA. This element is thought to be essential for viral replication and efficient packaging of avian infectious bronchitis virus (IBV) particles.
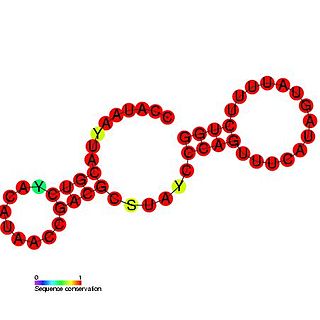
The Potato virus X cis-acting regulatory element is a cis-acting regulatory element found in the 3' UTR of the Potato virus X genome. This element has been found to be required for minus strand RNA accumulation and is essential for efficient viral replication.
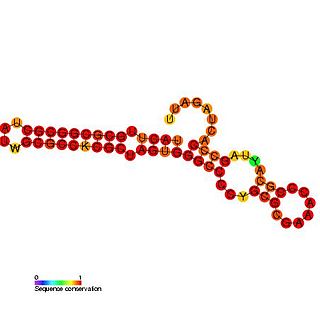
The Rubella virus 3′ cis-acting element RNA family represents a cis-acting element found at the 3′ UTR in the rubella virus. This family contains three conserved step loop structures. Calreticulin (CAL), which is known to bind calcium in most eukaryotic cells, is able to specifically bind to the first stem loop of this RNA. CAL binding is thought to be related to viral pathogenesis and in particular arthritis which occurs frequently in rubella infections in adults and is independent of viral viability. All stem loop structures are thought to be important for efficient viral replication and deletion of stem loop three is known to be lethal.
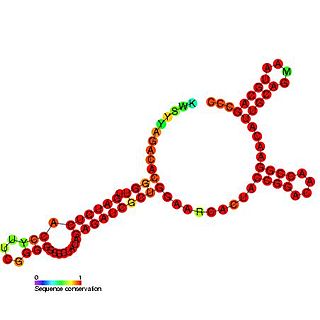
Tombusvirus 3′ UTR is an important cis-regulatory region of the Tombus virus genome.
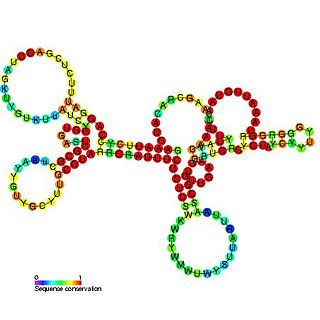
Tombusvirus 5′ UTR is an important cis-regulatory region of the Tombus virus genome.
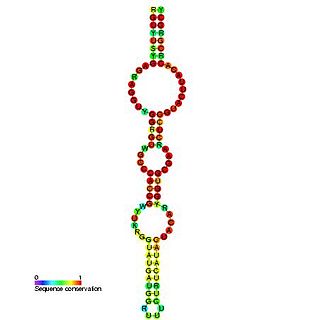
In virology, the tombusvirus internal replication element (IRE) is a segment of RNA located within the region coding for p92 polymerase. This element is essential for viral replication; specifically, it is thought to be required at an early stage of replication, such as template recruitment and/or replicase complex assembly.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 5′ end which is called the 5′ UTR. The 5′ UTR is responsible for important biological functions, such as viral replication, transcription and packaging. The 5′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 3′ end which is called the 3′ UTR. The 3′ UTR is responsible for important biological functions, such as viral replication. The 3′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.

Flavivirus 5' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.
Flavivirus 3' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.
A therapeutic interfering particle is an antiviral preparation that reduces the replication rate and pathogenesis of a particular viral infectious disease. A therapeutic interfering particle is typically a biological agent (i.e., nucleic acid) engineered from portions of the viral genome being targeted. Similar to Defective Interfering Particles (DIPs), the agent competes with the pathogen within an infected cell for critical viral replication resources, reducing the viral replication rate and resulting in reduced pathogenesis. But, in contrast to DIPs, TIPs are engineered to have an in vivo basic reproductive ratio (R0) that is greater than 1 (R0>1). The term "TIP" was first introduced in 2011 based on models of its mechanism-of-action from 2003. Given their unique R0>1 mechanism of action, TIPs exhibit high barriers to the evolution of antiviral resistance and are predicted to be resistance proof. Intervention with therapeutic interfering particles can be prophylactic (to prevent or ameliorate the effects of a future infection), or a single-administration therapeutic (to fight a disease that has already occurred, such as HIV or COVID-19). Synthetic DIPs that rely on stimulating innate antiviral immune responses (i.e., interferon) were proposed for influenza in 2008 and shown to protect mice to differing extents but are technically distinct from TIPs due to their alternate molecular mechanism of action which has not been predicted to have a similarly high barrier to resistance. Subsequent work tested the pre-clinical efficacy of TIPs against HIV, a synthetic DIP for SARS-CoV-2 (in vitro), and a TIP for SARS-CoV-2 (in vivo).














