
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). Averaged over multiple cell types in a given tissue, the quantity of mRNA is more than 10 times the quantity of ncRNA. The general preponderance of mRNA in cells is valid even though less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).
In molecular biology, the TATA box is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase.
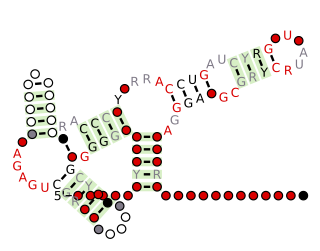
The TPP riboswitch, also known as the THI element and Thi-box riboswitch, is a highly conserved RNA secondary structure. It serves as a riboswitch that binds thiamine pyrophosphate (TPP) directly and modulate gene expression through a variety of mechanisms in archaea, bacteria and eukaryotes. TPP is the active form of thiamine (vitamin B1), an essential coenzyme synthesised by coupling of pyrimidine and thiazole moieties in bacteria. The THI element is an extension of a previously detected thiamin-regulatory element, the thi box, there is considerable variability in the predicted length and structures of the additional and facultative stem-loops represented in dark blue in the secondary structure diagram Analysis of operon structures has identified a large number of new candidate thiamin-regulated genes, mostly transporters, in various prokaryotic organisms. The x-ray crystal structure of the TPP riboswitch aptamer has been solved.

1-acyl-sn-glycerol-3-phosphate acyltransferase beta is an enzyme that in humans is encoded by the AGPAT2 gene.
Developmental noise is a concept within developmental biology in which the phenotype varies between individuals even though both the genotypes and the environmental factors are the same for all of them. Contributing factors include stochastic gene expression and other sources of cellular noise.

Alexander van Oudenaarden is a Dutch biophysicist and systems biologist. He is a leading researcher in stem cell biology, specialising in single cell techniques. In 2012 he started as director of the Hubrecht Institute and was awarded two times an ERC Advanced Grant, in 2012 and in 2017. He was awarded the Spinoza Prize in 2017.
Transcriptional bursting, also known as transcriptional pulsing, is a fundamental property of genes in which transcription from DNA to RNA can occur in "bursts" or "pulses", which has been observed in diverse organisms, from bacteria to mammals.

Robustness of a biological system is the persistence of a certain characteristic or trait in a system under perturbations or conditions of uncertainty. Robustness in development is known as canalization. According to the kind of perturbation involved, robustness can be classified as mutational, environmental, recombinational, or behavioral robustness etc. Robustness is achieved through the combination of many genetic and molecular mechanisms and can evolve by either direct or indirect selection. Several model systems have been developed to experimentally study robustness and its evolutionary consequences.
Cellular noise is random variability in quantities arising in cellular biology. For example, cells which are genetically identical, even within the same tissue, are often observed to have different expression levels of proteins, different sizes and structures. These apparently random differences can have important biological and medical consequences.
Johan Paulsson is a Swedish mathematician and systems biologist at Harvard Medical School. He is a leading researcher in systems biology and stochastic processes, specializing in stochasticity in gene networks and plasmid reproduction.

Synthetic biological circuits are an application of synthetic biology where biological parts inside a cell are designed to perform logical functions mimicking those observed in electronic circuits. The applications range from simply inducing production to adding a measurable element, like GFP, to an existing natural biological circuit, to implementing completely new systems of many parts.
intrinsic Noise Analyzer (iNA) is an open source software for studying reaction kinetics in living cells. The software analyzes mathematical models of intracellular reaction kinetics such as gene expression, regulatory networks or signaling pathways to quantify concentration fluctuations due to the random nature of chemical reactions.

CRISPR interference (CRISPRi) is a genetic perturbation technique that allows for sequence-specific repression of gene expression in prokaryotic and eukaryotic cells. It was first developed by Stanley Qi and colleagues in the laboratories of Wendell Lim, Adam Arkin, Jonathan Weissman, and Jennifer Doudna. Sequence-specific activation of gene expression refers to CRISPR activation (CRISPRa).
Single cell sequencing examines the sequence information from individual cells with optimized next-generation sequencing (NGS) technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear to be genetically clonal, but single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments.

In cell biology, single-cell variability occurs when individual cells in an otherwise similar population differ in shape, size, position in the cell cycle, or molecular-level characteristics. Such differences can be detected using modern single-cell analysis techniques. Investigation of variability within a population of cells contributes to understanding of developmental and pathological processes,
Leor S. Weinberger is an American virologist and quantitative biologist. He is credited with discovering the HIV virus latency circuit, which provided the first experimental evidence that stochastic fluctuations ('noise') in gene expression are used for cell fate decisions. He has also pioneered the concept of therapeutic interfering particles, or “TIPs”, which are resistance-proof antivirals. His TED talk on this novel antiviral approach 20 years in the making has been called a "highlight" of TED and received a standing ovation from the live audience.
State switching is a fundamental physiological process in which a cell/organism undergoes spontaneous, and potentially reversible, transitions between different phenotypes. Thus, the ability to switch states/phenotypes is a key feature of development and normal function of cells within most multicellular organisms that enables the cell to respond to various intrinsic and extrinsic cues and stimuli in a concerted fashion enabling them to ‘make’ appropriate cellular decisions. Although state switching is essential for normal functioning, the repertoire of phenotypes in a normal cell is albeit limited.
Perturb-seq refers to a high-throughput method of performing single cell RNA sequencing (scRNA-seq) on pooled genetic perturbation screens. Perturb-seq combines multiplexed CRISPR mediated gene inactivations with single cell RNA sequencing to assess comprehensive gene expression phenotypes for each perturbation. Inferring a gene’s function by applying genetic perturbations to knock down or knock out a gene and studying the resulting phenotype is known as reverse genetics. Perturb-seq is a reverse genetics approach that allows for the investigation of phenotypes at the level of the transcriptome, to elucidate gene functions in many cells, in a massively parallel fashion.