
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein, or to sequence or quantify mRNA molecules using DNA based methods. cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq.

Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.
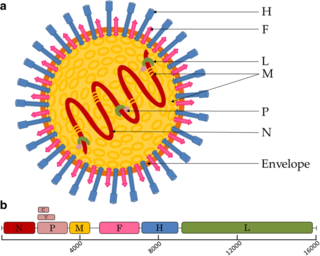
Paramyxoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates serve as natural hosts. Diseases associated with this family include measles, mumps, and respiratory tract infections. The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily.

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Cauliflower mosaic virus (CaMV) is a member of the genus Caulimovirus, one of the six genera in the family Caulimoviridae, which are pararetroviruses that infect plants. Pararetroviruses replicate through reverse transcription just like retroviruses, but the viral particles contain DNA instead of RNA.

Tombusviridae is a family of single-stranded positive sense RNA plant viruses. There are three subfamilies, 17 genera, and 95 species in this family. The name is derived from Tomato bushy stunt virus (TBSV).
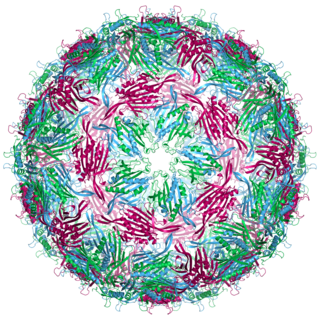
Bacteriophage MS2, commonly called MS2, is an icosahedral, positive-sense single-stranded RNA virus that infects the bacterium Escherichia coli and other members of the Enterobacteriaceae. MS2 is a member of a family of closely related bacterial viruses that includes bacteriophage f2, bacteriophage Qβ, R17, and GA.

Sindbis virus (SINV) is a member of the Togaviridae family, in the Alphavirus genus. The virus was first isolated in 1952 in Cairo, Egypt. The virus is transmitted by mosquitoes. SINV is linked to Pogosta disease (Finland), Ockelbo disease (Sweden) and Karelian fever (Russia). In humans, the symptoms include arthralgia, rash and malaise. Sindbis virus is widely and continuously found in insects and vertebrates in Eurasia, Africa, and Oceania. Clinical infection and disease in humans however has almost only been reported from Northern Europe, where SINV is endemic and where large outbreaks occur intermittently. Cases are occasionally reported in Australia, China, and South Africa.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase.

Alfalfa mosaic virus (AMV), also known as Lucerne mosaic virus or Potato calico virus, is a worldwide distributed phytopathogen that can lead to necrosis and yellow mosaics on a large variety of plant species, including commercially important crops. It is the only Alfamovirus of the family Bromoviridae. In 1931 Weimer J.L. was the first to report AMV in alfalfa. Transmission of the virus occurs mainly by some aphids, by seeds or by pollen to the seed.

The equine arteritis virus leader transcription-regulating sequence hairpin (LTH) is as RNA element that is thought to be a key structural element in discontinuous subgenomic RNA synthesis and is critical for leader transcription-regulating sequences (TRS) function. Similar structures have been predicted in other arteriviruses and coronaviruses.
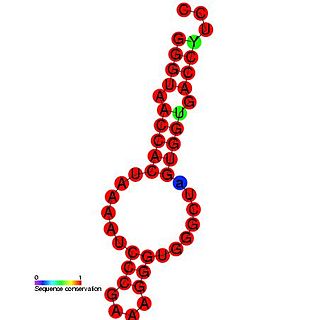
The TCV hairpin 5 (H5) is an RNA element found in the turnip crinkle virus. This RNA element is composed of a stem-loop that contains a large symmetrical internal loop (LSL). H5 can repress minus-strand synthesis when the 3' side of the LSL pairs with the 4 bases at the 3'-terminus of the RNA(GCCC-OH).

The Upstream pseudoknot (UPSK) domain is an RNA element found in the turnip yellow mosaic virus, beet virus Q, barley stripe mosaic virus and tobacco mosaic virus, which is thought to be needed for efficient transcription. Disruption of the pseudoknot structure gives rise to a 50% drop in transcription efficiency. This element acts in conjunction with the Tymovirus/Pomovirus tRNA-like 3' UTR element to enhance translation.
Turnip crinkle virus (TCV) is a plant pathogenic virus of the family Tombusviridae. It was first isolated from turnip.

Transfer RNA-like structures are RNA sequences, which have a similar tertiary structure to tRNA; they frequently contain a pseudoknot close to the 3' end. The presence of tRNA-like structures has been demonstrated in many plant virus RNA genomes. These tRNA-like structures are linked to regulation of plant virus replication.
In molecular biology, Turnip crinkle virus (TCV) hairpin H4 is an RNA hairpin found at the 3' end of the Turnip crinkle virus (TCV) genome.
In molecular biology, a cap-independent translation element is an RNA sequence found in the 3'UTR of many RNA plant viruses.

Ground squirrel hepatitis virus, abbreviated GSHV, is a partially double-stranded DNA virus that is closely related to human Hepatitis B virus (HBV) and Woodchuck hepatitis virus (WHV). It is a member of the family of viruses Hepadnaviridae and the genus Orthohepadnavirus. Like the other members of its family, GSHV has high degree of species and tissue specificity. It was discovered in Beechey ground squirrels, Spermophilus beecheyi, but also infects Arctic ground squirrels, Spermophilus parryi. Commonalities between GSHV and HBV include morphology, DNA polymerase activity in genome repair, cross-reacting viral antigens, and the resulting persistent infection with viral antigen in the blood (antigenemia). As a result, GSHV is used as an experimental model for HBV.
Rolling hairpin replication (RHR) is a unidirectional, strand displacement form of DNA replication used by parvoviruses, a group of viruses that constitute the family Parvoviridae. Parvoviruses have linear, single-stranded DNA (ssDNA) genomes in which the coding portion of the genome is flanked by telomeres at each end that form hairpin loops. During RHR, these hairpin loops repeatedly unfold and refold to change the direction of DNA replication so that replication progresses in a continuous manner back and forth across the genome. RHR is initiated and terminated by an endonuclease encoded by parvoviruses that is variously called NS1 or Rep, and RHR is similar to rolling circle replication, which is used by ssDNA viruses that have circular genomes.













