
Poliovirus, the causative agent of polio, is a serotype of the species Enterovirus C, in the family of Picornaviridae. There are three poliovirus serotypes: types 1, 2, and 3.
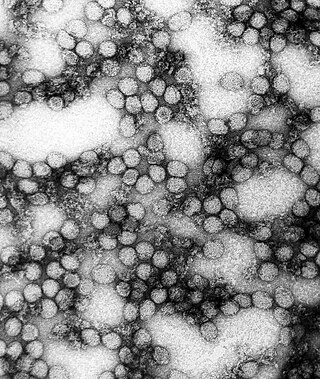
Flavivirus, renamed Orthoflavivirus in 2023, is a genus of positive-strand RNA viruses in the family Flaviviridae. The genus includes the West Nile virus, dengue virus, tick-borne encephalitis virus, yellow fever virus, Zika virus and several other viruses which may cause encephalitis, as well as insect-specific flaviviruses (ISFs) such as cell fusing agent virus (CFAV), Palm Creek virus (PCV), and Parramatta River virus (PaRV). While dual-host flaviviruses can infect vertebrates as well as arthropods, insect-specific flaviviruses are restricted to their competent arthropods. The means by which flaviviruses establish persistent infection in their competent vectors and cause disease in humans depends upon several virus-host interactions, including the intricate interplay between flavivirus-encoded immune antagonists and the host antiviral innate immune effector molecules.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.

Sindbis virus (SINV) is a member of the Togaviridae family, in the Alphavirus genus. The virus was first isolated in 1952 in Cairo, Egypt. The virus is transmitted by mosquitoes. SINV is linked to Pogosta disease (Finland), Ockelbo disease (Sweden) and Karelian fever (Russia). In humans, the symptoms include arthralgia, rash and malaise. Sindbis virus is widely and continuously found in insects and vertebrates in Eurasia, Africa, and Oceania. Clinical infection and disease in humans however has almost only been reported from Northern Europe, where SINV is endemic and where large outbreaks occur intermittently. Cases are occasionally reported in Australia, China, and South Africa.

The Alfalfa mosaic virus RNA 1 5′ UTR stem-loop represents a putative stem-loop structure found in the 5′ UTR in RNA 1 of alfalfa mosaic virus. RNA 1 is responsible for encoding the viral replicase protein P1. This family is required for negative strand RNA synthesis in the alfalfa mosaic virus and may also be involved in positive strand RNA synthesis.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Betacoronavirus. It has an important role in regulating the packaging of the viral genome into the capsid. As part of the viral life cycle, within the infected cell, the viral genome becomes associated with viral proteins and assembles into new infective progeny viruses. This process is called packaging and is vital for viral replication.
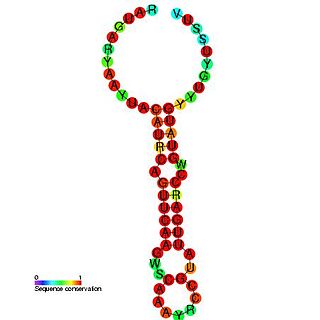
Enterovirus cis-acting replication element is a small RNA hairpin in the coding region of protein 2C as the site in PV1(M) RNA that is used as the primary template for the in vitro uridylylation. The first step in the replication of the plus-stranded poliovirus RNA is the synthesis of a complementary minus strand. This process is initiated by the covalent attachment of uridine monophosphate (UMP) to the terminal protein VPg, yielding VPgpU and VPgpUpU.
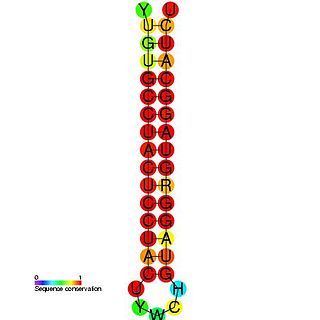
The Hepatitis C stem-loop IV is part of a putative RNA element found in the NS5B coding region. This element along with stem-loop VII, is important for colony formation, though its exact function and mechanism are unknown.

The hepatitis C virus 3′X element is an RNA element which contains three stem-loop structures that are essential for replication.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.
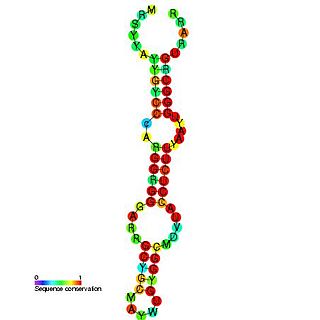
Hepatitis C virus stem-loop VII is a regulatory element found in the coding region of the RNA-dependent RNA polymerase gene, NS5B. Similarly to stem-loop IV, the stem-loop structure is important for colony formation, though its exact function and mechanism are unknown.

The retroviral psi packaging element, also known as the Ψ RNA packaging signal, is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.
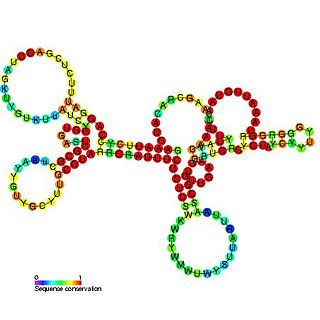
Tombusvirus 5′ UTR is an important cis-regulatory region of the Tombus virus genome.

The turnip crinkle virus (TCV) core promoter hairpin (Pr) is an RNA element located in the 3' UTR of the viral genome that is required for minus strand RNA synthesis. The picture shown is not the TCV core promoter, but an upstream hairpin that is also required for replication of the virus.

The Upstream pseudoknot (UPSK) domain is an RNA element found in the turnip yellow mosaic virus, beet virus Q, barley stripe mosaic virus and tobacco mosaic virus, which is thought to be needed for efficient transcription. Disruption of the pseudoknot structure gives rise to a 50% drop in transcription efficiency. This element acts in conjunction with the Tymovirus/Pomovirus tRNA-like 3' UTR element to enhance translation.
Turnip crinkle virus (TCV) is a plant pathogenic virus of the family Tombusviridae. It was first isolated from turnip.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.
In molecular biology, Turnip crinkle virus (TCV) hairpin H4 is an RNA hairpin found at the 3' end of the Turnip crinkle virus (TCV) genome.

Flavivirus 5' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.

















