Related Research Articles

A pilus is a hair-like appendage found on the surface of many bacteria and archaea. The terms pilus and fimbria can be used interchangeably, although some researchers reserve the term pilus for the appendage required for bacterial conjugation. All pili in the latter sense are primarily composed of pilin proteins, which are oligomeric.

Neisseria gonorrhoeae, also known as gonococcus (singular), or gonococci (plural), is a species of Gram-negative diplococci bacteria isolated by Albert Neisser in 1879. It causes the sexually transmitted genitourinary infection gonorrhea as well as other forms of gonococcal disease including disseminated gonococcemia, septic arthritis, and gonococcal ophthalmia neonatorum.

Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.

Genetic recombination is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be passed on from the parents to the offspring. Most recombination is naturally occurring.

Neisseria is a large genus of bacteria that colonize the mucosal surfaces of many animals. Of the 11 species that colonize humans, only two are pathogens, N. meningitidis and N. gonorrhoeae. Most gonococcal infections are asymptomatic and self-resolving, and epidemic strains of the meningococcus may be carried in >95% of a population where systemic disease occurs at <1% prevalence.
The restriction modification system is found in bacteria and other prokaryotic organisms, and provides a defense against foreign DNA, such as that borne by bacteriophages.
Gene duplication is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. Gene duplications can arise as products of several types of errors in DNA replication and repair machinery as well as through fortuitous capture by selfish genetic elements. Common sources of gene duplications include ectopic recombination, retrotransposition event, aneuploidy, polyploidy, and replication slippage.

In molecular biology and genetics, transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous genetic material from its surroundings through the cell membrane(s). For transformation to take place, the recipient bacterium must be in a state of competence, which might occur in nature as a time-limited response to environmental conditions such as starvation and cell density, and may also be induced in a laboratory.
Multilocus sequence typing (MLST) is a technique in molecular biology for the typing of multiple loci, using DNA sequences of internal fragments of multiple housekeeping genes to characterize isolates of microbial species.

Neisseria meningitidis, often referred to as meningococcus, is a Gram-negative bacterium that can cause meningitis and other forms of meningococcal disease such as meningococcemia, a life-threatening sepsis. The bacterium is referred to as a coccus because it is round, and more specifically a diplococcus because of its tendency to form pairs.
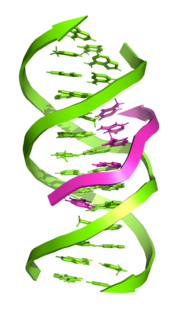
Triple-stranded DNA is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a B-form DNA double helix by forming Hoogsteen base pairs or reversed Hoogsteen hydrogen bonds.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.

In microbiology, genetics, cell biology, and molecular biology, competence is the ability of a cell to alter its genetics by taking up extracellular ("naked") DNA from its environment in the process called transformation. Competence may be differentiated between natural competence, a genetically specified ability of bacteria which is thought to occur under natural conditions as well as in the laboratory, and induced or artificial competence, which arises when cells in laboratory cultures are treated to make them transiently permeable to DNA. Competence allows for rapid adaptation and DNA repair of the cell. This article primarily deals with natural competence in bacteria, although information about artificial competence is also provided.
Antigenic variation or antigenic alteration refers to the mechanism by which an infectious agent such as a protozoan, bacterium or virus alters the proteins or carbohydrates on its surface and thus avoids a host immune response, making it one of the mechanisms of antigenic escape. It is related to phase variation. Antigenic variation not only enables the pathogen to avoid the immune response in its current host, but also allows re-infection of previously infected hosts. Immunity to re-infection is based on recognition of the antigens carried by the pathogen, which are "remembered" by the acquired immune response. If the pathogen's dominant antigen can be altered, the pathogen can then evade the host's acquired immune system. Antigenic variation can occur by altering a variety of surface molecules including proteins and carbohydrates. Antigenic variation can result from gene conversion, site-specific DNA inversions, hypermutation, or recombination of sequence cassettes. The result is that even a clonal population of pathogens expresses a heterogeneous phenotype. Many of the proteins known to show antigenic or phase variation are related to virulence.
Pilin refers to a class of fibrous proteins that are found in pilus structures in bacteria. These structures can be used for the exchange of genetic material, or as a cell adhesion mechanism. Although not all bacteria have pili or fimbriae, bacterial pathogens often use their fimbriae to attach to host cells. In Gram-negative bacteria, where pili are more common, individual pilin molecules are linked by noncovalent protein-protein interactions, while Gram-positive bacteria often have polymerized LPXTG pilin.
Fosmids are similar to cosmids but are based on the bacterial F-plasmid. The cloning vector is limited, as a host can only contain one fosmid molecule. Fosmids can hold DNA inserts of up to 40 kb in size; often the source of the insert is random genomic DNA. A fosmid library is prepared by extracting the genomic DNA from the target organism and cloning it into the fosmid vector. The ligation mix is then packaged into phage particles and the DNA is transfected into the bacterial host. Bacterial clones propagate the fosmid library. The low copy number offers higher stability than vectors with relatively higher copy numbers, including cosmids. Fosmids may be useful for constructing stable libraries from complex genomes. Fosmids have high structural stability and have been found to maintain human DNA effectively even after 100 generations of bacterial growth. Fosmid clones were used to help assess the accuracy of the Public Human Genome Sequence.

Pasteurella multocida is a Gram-negative, nonmotile, penicillin-sensitive coccobacillus of the family Pasteurellaceae. Strains of the species are currently classified into five serogroups based on capsular composition and 16 somatic serovars (1–16). P. multocida is the cause of a range of diseases in mammals and birds, including fowl cholera in poultry, atrophic rhinitis in pigs, and bovine hemorrhagic septicemia in cattle and buffalo. It can also cause a zoonotic infection in humans, which typically is a result of bites or scratches from domestic pets. Many mammals and birds harbor it as part of their normal respiratory microbiota.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.
Bacterial recombination is a type of genetic recombination in bacteria characterized by DNA transfer from one organism called donor to another organism as recipient. This process occurs in three main ways:

In bacteria, phasevarions mediate a coordinated change in the expression of multiple genes or proteins. This occurs via phase variation of a single DNA methyltransferase. Phase variation of methyltransferase expression results in differential methylation throughout the bacterial genome, leading to variable expression of multiple genes through epigenetic mechanisms.
References
- ↑ Frye SA, Nilsen M, Tønjum T, Ambur OH (2013). "Dialects of the DNA uptake sequence in Neisseriaceae". PLOS Genet. 9 (4): 3065–70. doi:10.1371/journal.pgen.1003458. PMC 3630211 . PMID 23637627.
- ↑ Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V (2013). "Specific DNA recognition mediated by a type IV pilin". Proc. Natl. Acad. Sci. U.S.A. 110 (8): 3065–70. Bibcode:2013PNAS..110.3065C. doi: 10.1073/pnas.1218832110 . PMC 3581936 . PMID 23386723.
- ↑ Davidsen T, Rødland EA, Lagesen K, Seeberg E, Rognes T, Tønjum T (2004). "Biased distribution of DNA uptake sequences towards genome maintenance genes". Nucleic Acids Res. 32 (3): 1050–8. doi:10.1093/nar/gkh255. PMC 373393 . PMID 14960717.