Related Research Articles
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations. The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB.

Social network analysis (SNA) is the process of investigating social structures through the use of networks and graph theory. It characterizes networked structures in terms of nodes and the ties, edges, or links that connect them. Examples of social structures commonly visualized through social network analysis include social media networks, meme spread, information circulation, friendship and acquaintance networks, peer learner networks, business networks, knowledge networks, difficult working relationships, collaboration graphs, kinship, disease transmission, and sexual relationships. These networks are often visualized through sociograms in which nodes are represented as points and ties are represented as lines. These visualizations provide a means of qualitatively assessing networks by varying the visual representation of their nodes and edges to reflect attributes of interest.

Structural bioinformatics is the branch of bioinformatics that is related to the analysis and prediction of the three-dimensional structure of biological macromolecules such as proteins, RNA, and DNA. It deals with generalizations about macromolecular 3D structures such as comparisons of overall folds and local motifs, principles of molecular folding, evolution, binding interactions, and structure/function relationships, working both from experimentally solved structures and from computational models. The term structural has the same meaning as in structural biology, and structural bioinformatics can be seen as a part of computational structural biology. The main objective of structural bioinformatics is the creation of new methods of analysing and manipulating biological macromolecular data in order to solve problems in biology and generate new knowledge.
XLispStat is a statistical scientific package based on the XLISP language.

Data analysis is the process of inspecting, cleansing, transforming, and modeling data with the goal of discovering useful information, informing conclusions, and supporting decision-making. Data analysis has multiple facets and approaches, encompassing diverse techniques under a variety of names, and is used in different business, science, and social science domains. In today's business world, data analysis plays a role in making decisions more scientific and helping businesses operate more effectively.
CAD data exchange is a method of drawing data exchange used to translate between different computer-aided design (CAD) authoring systems or between CAD and other downstream CAx systems.
Graph Modeling Language (GML) is a hierarchical ASCII-based file format for describing graphs. It has been also named Graph Meta Language.
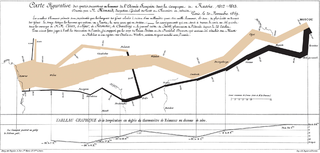
Data and information visualization is the practice of designing and creating easy-to-communicate and easy-to-understand graphic or visual representations of a large amount of complex quantitative and qualitative data and information with the help of static, dynamic or interactive visual items. Typically based on data and information collected from a certain domain of expertise, these visualizations are intended for a broader audience to help them visually explore and discover, quickly understand, interpret and gain important insights into otherwise difficult-to-identify structures, relationships, correlations, local and global patterns, trends, variations, constancy, clusters, outliers and unusual groupings within data. When intended for the general public to convey a concise version of known, specific information in a clear and engaging manner, it is typically called information graphics.
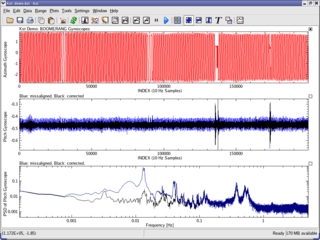
Kst is a plotting and data viewing program. It is a general purpose plotting software program that evolved out of a need to visualize and analyze astronomical data, but has also found subsequent use in the real time display of graphical information. Kst is a KDE application and is freely available for anyone to download and use under the terms of the GPL. It is noted for being able to graph real-time data acquisition.
The completion of the human genome sequencing in the early 2000s was a turning point in genomics research. Scientists have conducted series of research into the activities of genes and the genome as a whole. The human genome contains around 3 billion base pairs nucleotide, and the huge quantity of data created necessitates the development of an accessible tool to explore and interpret this information in order to investigate the genetic basis of disease, evolution, and biological processes. The field of genomics has continued to grow, with new sequencing technologies and computational tool making it easier to study the genome.

UCSF Chimera is an extensible program for interactive visualization and analysis of molecular structures and related data, including density maps, supramolecular assemblies, sequence alignments, docking results, trajectories, and conformational ensembles. High-quality images and movies can be created. Chimera includes complete documentation and can be downloaded free of charge for noncommercial use.

Tecplot is the name of a family of visualization & analysis software tools developed by American company Tecplot, Inc., which is headquartered in Bellevue, Washington. The firm was formerly operated as Amtec Engineering. In 2016, the firm was acquired by Vela Software, an operating group of Constellation Software, Inc. (TSX:CSU).

UGENE is computer software for bioinformatics. It works on personal computer operating systems such as Windows, macOS, or Linux. It is released as free and open-source software, under a GNU General Public License (GPL) version 2.

Voreen is an open-source volume visualization library and development platform. Through the use of GPU-based volume rendering techniques it allows high frame rates on standard graphics hardware to support interactive volume exploration.

ELKI is a data mining software framework developed for use in research and teaching. It was originally at the database systems research unit of Professor Hans-Peter Kriegel at the Ludwig Maximilian University of Munich, Germany, and now continued at the Technical University of Dortmund, Germany. It aims at allowing the development and evaluation of advanced data mining algorithms and their interaction with database index structures.

Wolfgang Hackbusch is a German mathematician, known for his pioneering research in multigrid methods and later hierarchical matrices, a concept generalizing the fast multipole method. He was a professor at the University of Kiel and is currently one of the directors of the Max Planck Institute for Mathematics in the Sciences in Leipzig.

MeVisLab is a cross-platform application framework for medical image processing and scientific visualization. It includes advanced algorithms for image registration, segmentation, and quantitative morphological and functional image analysis. An IDE for graphical programming and rapid user interface prototyping is available.

Gephi is an open-source network analysis and visualization software package written in Java on the NetBeans platform.

Amira is a software platform for 3D and 4D data visualization, processing, and analysis. It is being actively developed by Thermo Fisher Scientific in collaboration with the Zuse Institute Berlin (ZIB), and commercially distributed by Thermo Fisher Scientific.

NodeXL is a network analysis and visualization software package for Microsoft Excel 2007/2010/2013/2016. The package is similar to other network visualization tools such as Pajek, UCINet, and Gephi. It is widely applied in ring, mapping of vertex and edge, and customizable visual attributes and tags. NodeXL enables researchers to undertake social network analysis work metrics such as centrality, degree, and clustering, as well as monitor relational data and describe the overall relational network structure. When applied to Twitter data analysis, it showed the total network of all users participating in public discussion and its internal structure through data mining. It allows social Network analysis (SNA) to emphasize the relationships rather than the isolated individuals or organizations, allowing interested parties to investigate the two-way dialogue between organizations and the public. SNA also provides a flexible measurement system and parameter selection to confirm the influential nodes in the network, such as in-degree and out-degree centrality. The software contains network visualization, social network analysis features, access to social media network data importers, advanced network metrics, and automation.
References
- K. Gruchalla, M. Rast, E. Bradley, J. Clyne, P. Mininni, Visualization-Driven Structural and Statistical Analysis of Turbulent Flows, Procs. IDA 2009, Lecture Notes in Computer Science 5772, pp 321-332, Springer, 2009 doi : 10.1007/978-3-642-03915-7_28 ISBN 978-3-642-03914-0