Related Research Articles
Endoreduplication is replication of the nuclear genome in the absence of mitosis, which leads to elevated nuclear gene content and polyploidy. Endoreplication can be understood simply as a variant form of the mitotic cell cycle (G1-S-G2-M) in which mitosis is circumvented entirely, due to modulation of cyclin-dependent kinase (CDK) activity. Examples of endoreplication characterized in arthropod, mammalian, and plant species suggest that it is a universal developmental mechanism responsible for the differentiation and morphogenesis of cell types that fulfill an array of biological functions. While endoreplication is often limited to specific cell types in animals, it is considerably more widespread in plants, such that polyploidy can be detected in the majority of plant tissues.
Florigen is the hypothesized hormone-like molecule responsible for controlling and/or triggering flowering in plants. Florigen is produced in the leaves, and acts in the shoot apical meristem of buds and growing tips. It is known to be graft-transmissible, and even functions between species. Florigen has been found identical to the transcription factor FLOWERING LOCUS T (FT).
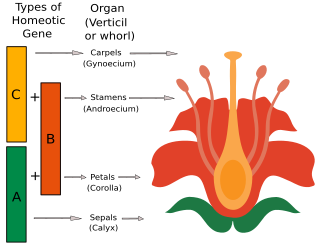
The ABC model of flower development is a scientific model of the process by which flowering plants produce a pattern of gene expression in meristems that leads to the appearance of an organ oriented towards sexual reproduction, a flower. There are three physiological developments that must occur in order for this to take place: firstly, the plant must pass from sexual immaturity into a sexually mature state ; secondly, the transformation of the apical meristem's function from a vegetative meristem into a floral meristem or inflorescence; and finally the growth of the flower's individual organs. The latter phase has been modelled using the ABC model, which aims to describe the biological basis of the process from the perspective of molecular and developmental genetics.

MicroRNA (miRNA) precursor miR156 is a family of plant non-coding RNA. This microRNA has now been predicted or experimentally confirmed in a range of plant species. Animal miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. miR156 functions in the induction of flowering by suppressing the transcripts of SQUAMOSA-PROMOTER BINDING LIKE (SPL) transcription factors gene family. It was suggested that the loading into ARGONAUTE1 and ARGONAUTE5 is required for miR156 functionality in Arabidopsis thaliana. In plants the precursor sequences may be longer, and the carpel factory (caf) enzyme appears to be involved in processing. In this case the mature sequence comes from the 5' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA.
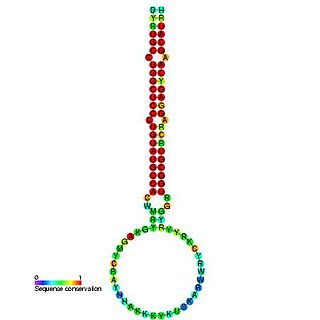
In molecular biology, mir-160 is a microRNA that has been predicted or experimentally confirmed in a range of plant species including Arabidopsis thaliana and Oryza sativa (rice). miR-160 is predicted to bind complementary sites in the untranslated regions of auxin response factor genes to regulate their expression. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.

The mir-172 microRNA is thought to target mRNAs coding for APETALA2-like transcription factors. It has been verified experimentally in the model plant, Arabidopsis thaliana. The mature sequence is excised from the 3' arm of the hairpin.

mir-395 is a non-coding RNA called a microRNA that was identified in both Arabidopsis thaliana and Oryza sativa computationally and was later experimentally verified. mir-395 is thought to target mRNAs coding for ATP sulphurylases. The mature sequence is excised from the 3' arm of the hairpin.
Evolutionary developmental biology (evo-devo) is the study of developmental programs and patterns from an evolutionary perspective. It seeks to understand the various influences shaping the form and nature of life on the planet. Evo-devo arose as a separate branch of science rather recently. An early sign of this occurred in 1999.
Trans-acting siRNA are a class of small interfering RNA (siRNA) that repress gene expression through post-transcriptional gene silencing in land plants. Precursor transcripts from TAS loci are polyadenylated and converted to double-stranded RNA, and are then processed into 21-nucleotide-long RNA duplexes with overhangs. These segments are incorporated into an RNA-induced silencing complex (RISC) and direct the sequence-specific cleavage of target mRNA. Ta-siRNAs are classified as siRNA because they arise from double-stranded RNA (dsRNA).
In molecular biology mir-390 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-396 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-408 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-824 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-398 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
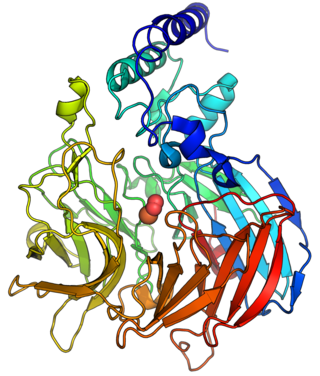
9-cis-epoxycarotenoid dioxygenase (EC 1.13.11.51, nine-cis-epoxycarotenoid dioxygenase, NCED, AtNCED3, PvNCED1, VP14) is an enzyme in the biosynthesis of abscisic acid (ABA), with systematic name 9-cis-epoxycarotenoid 11,12-dioxygenase. This enzyme catalyses the following chemical reaction

Liam Dolan is a Senior Group Leader at the Gregor Mendel Institute of Molecular Plant Biology (GMI) of the Austrian Academy of Sciences, the Sherardian Professor of Botany in the Department of Biology at the University of Oxford and a Fellow of Magdalen College, Oxford.
LUX or Phytoclock1 (PCL1) is a gene that codes for LUX ARRHYTHMO, a protein necessary for circadian rhythms in Arabidopsis thaliana. LUX protein associates with Early Flowering 3 (ELF3) and Early Flowering 4 (ELF4) to form the Evening Complex (EC), a core component of the Arabidopsis repressilator model of the plant circadian clock. The LUX protein functions as a transcription factor that negatively regulates Pseudo-Response Regulator 9 (PRR9), a core gene of the Midday Complex, another component of the Arabidopsis repressilator model. LUX is also associated with circadian control of hypocotyl growth factor genes PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PHYTOCHROME INTERACTING FACTOR 5 (PIF5).
Arabidopsis thaliana is a first class model organism and the single most important species for fundamental research in plant molecular genetics.

Sarah Hake is an American plant developmental biologist who directs the USDA's Plant Gene Expression Center in Albany, CA. In 2009 she was elected a fellow of the American Association for the Advancement of Science and elected member of the National Academy of Sciences.

Paula McSteen is a scientist known for her research on plant genetics. In 2020 she was elected a fellow of the American Association for the Advancement of Science.
References
- 1 2 Poethig, R. S. (2010). "The past, present, and future of vegetative phase change". Plant Physiology. 154 (2): 541–544. doi:10.1104/pp.110.161620. PMC 2949024 . PMID 20921181.
- ↑ Chuck, G. S.; Tobias, C.; Sun, L.; Kraemer, F.; Li, C.; Dibble, D.; Arora, R.; Bragg, J. N.; Vogel, J. P.; Singh, S.; Simmons, B. A.; Pauly, M.; Hake, S. (2011). "Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass". Proceedings of the National Academy of Sciences. 108 (42): 17550–17555. doi: 10.1073/pnas.1113971108 . PMC 3198312 . PMID 21987797.
- ↑ Telfer, A.; Bollman, K. M.; Poethig, R. S. (1997). "Phase change and the regulation of trichome distribution in Arabidopsis thaliana". Development. 124 (3): 645–654. doi:10.1242/dev.124.3.645. PMID 9043079.
- ↑ Wu, G.; Poethig, R. S. (2006). "Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3". Development. 133 (18): 3539–3547. doi:10.1242/dev.02521. PMC 1610107 . PMID 16914499.
- ↑ Chuck, G.; Cigan, A. M.; Saeteurn, K.; Hake, S. (2007). "The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA". Nature Genetics. 39 (4): 544–549. doi:10.1038/ng2001. PMID 17369828. S2CID 20014922.