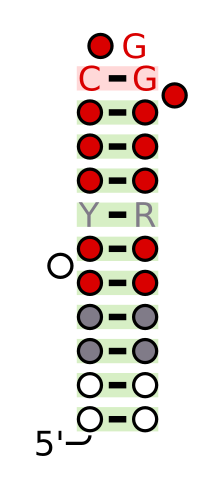
The signal recognition particle (SRP) is an abundant, cytosolic, universally conserved ribonucleoprotein that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and the plasma membrane in prokaryotes.

A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple catalytic domains are found in a single polypeptide chain.

Pleckstrin homology domain or (PHIP) is a protein domain of approximately 120 amino acids that occurs in a wide range of proteins involved in intracellular signaling or as constituents of the cytoskeleton.
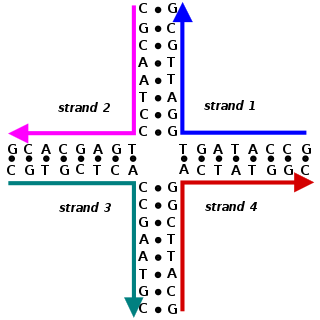
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after Robin Holliday, the molecular biologist who proposed its existence in 1964.
The MADS box is a conserved sequence motif. The genes which contain this motif are called the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC[A/T]6GG termed the CArG-box. MADS-domain proteins are generally transcription factors. The length of the MADS-box reported by various researchers varies somewhat, but typical lengths are in the range of 168 to 180 base pairs, i.e. the encoded MADS domain has a length of 56 to 60 amino acids. There is evidence that the MADS domain evolved from a sequence stretch of a type II topoisomerase in a common ancestor of all extant eukaryotes.

The exosome complex is a multi-protein intracellular complex capable of degrading various types of RNA molecules. Exosome complexes are found in both eukaryotic cells and archaea, while in bacteria a simpler complex called the degradosome carries out similar functions.

Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.
Cell wall protein 2 (CWP2) is a cell wall protein, produced by Saccharomyces cerevisiae and Saccharomyces pastorianus. It occurs throughout the cell wall and has close homology with the CWP1 gene.

U2 spliceosomal snRNAs are a species of small nuclear RNA (snRNA) molecules found in the major spliceosomal (Sm) machinery of virtually all eukaryotic organisms. In vivo, U2 snRNA along with its associated polypeptides assemble to produce the U2 small nuclear ribonucleoprotein (snRNP), an essential component of the major spliceosomal complex. The major spliceosomal-splicing pathway is occasionally referred to as U2 dependent, based on a class of Sm intron—found in mRNA primary transcripts—that are recognized exclusively by the U2 snRNP during early stages of spliceosomal assembly. In addition to U2 dependent intron recognition, U2 snRNA has been theorized to serve a catalytic role in the chemistry of pre-RNA splicing as well. Similar to ribosomal RNAs (rRNAs), Sm snRNAs must mediate both RNA:RNA and RNA:protein contacts and hence have evolved specialized, highly conserved, primary and secondary structural elements to facilitate these types of interactions.

U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes.

The signal recognition particle RNA, is part of the signal recognition particle (SRP) ribonucleoprotein complex. SRP recognizes the signal peptide and binds to the ribosome, halting protein synthesis. SRP-receptor is a protein that is embedded in a membrane, and which contains a transmembrane pore. When the SRP-ribosome complex binds to SRP-receptor, SRP releases the ribosome and drifts away. The ribosome resumes protein synthesis, but now the protein is moving through the SRP-receptor transmembrane pore.

TRAMP complex is a multiprotein, heterotrimeric complex having distributive polyadenylation activity and identifies wide varieties of RNAs produced by polymerases. It was originally discovered in Saccharomycescerevisiae by LaCava et al., Vanacova et al. and Wyers et al. in 2005.
Tyrosine—tRNA ligase, also known as tyrosyl-tRNA synthetase is an enzyme that is encoded by the gene YARS. Tyrosine—tRNA ligase catalyzes the chemical reaction

U1 small nuclear ribonucleoprotein A is a protein that in humans is encoded by the SNRPA gene.

NHP2-like protein 1 is a protein that in humans is encoded by the SNU13 gene.
Z-DNA binding protein 1, also known as Zuotin, is a Saccharomyces cerevisiae yeast gene.

In molecular biology, the protein domain Sterile alpha motif is a putative protein interaction module present in a wide variety of proteins involved in many biological processes. The SAM domain that spreads over around 70 residues is found in diverse eukaryotic organisms. SAM domains have been shown to homo- and hetero-oligomerise, forming multiple self-association architectures and also binding to various non-SAM domain-containing proteins, nevertheless with a low affinity constant.

In molecular biology, the single-domain protein SUI1 is a translation initiation factor often found in the fungus, Saccharomyces cerevisiae but it is also found in other eukaryotes and prokaryotes as well as archaea. It is otherwise known as Eukaryotic translation initiation factor 1 (eIF1) in eukaryotes or YciH in bacteria.
The transactivation domain or trans-activating domain (TAD) is a transcription factor scaffold domain which contains binding sites for other proteins such as transcription coregulators. These binding sites are frequently referred to as activation functions (AFs). TADs are named after their amino acid composition. These amino acids are either essential for the activity or simply the most abundant in the TAD. Transactivation by the Gal4 transcription factor is mediated by acidic amino acids, whereas hydrophobic residues in Gcn4 play a similar role. Hence, the TADs in Gal4 and Gcn4 are referred to as acidic or hydrophobic, respectively.
The Gal4 transcription factor is a positive regulator of gene expression of galactose-induced genes. This protein represents a large fungal family of transcription factors, Gal4 family, which includes over 50 members in the yeast Saccharomyces cerevisiae e.g. Oaf1, Pip2, Pdr1, Pdr3, Leu3.