
An alpha helix is a sequence of amino acids in a protein that are twisted into a coil.
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs may consist of one or several alpha-helices or a transmembrane beta barrel. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in size and hydrophobicity; they may adopt organelle-specific properties.

Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane. Peripheral membrane proteins are transiently associated with the cell membrane.

A transmembrane protein is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.

Peripheral membrane proteins, or extrinsic membrane proteins, are membrane proteins that adhere only temporarily to the biological membrane with which they are associated. These proteins attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins.
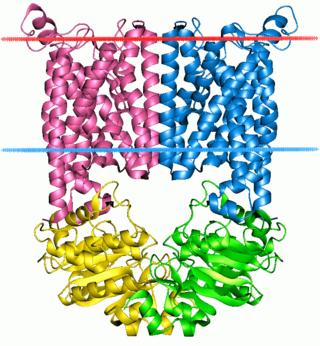
The ATP-binding cassette transporters are a transport system superfamily that is one of the largest and possibly one of the oldest gene families. It is represented in all extant phyla, from prokaryotes to humans. ABC transporters belong to translocases.

SNARE proteins – "SNAPREceptors" – are a large protein family consisting of at least 24 members in yeasts, more than 60 members in mammalian cells, and some numbers in plants. The primary role of SNARE proteins is to mediate the fusion of vesicles with the target membrane; this notably mediates exocytosis, but can also mediate the fusion of vesicles with membrane-bound compartments. The best studied SNAREs are those that mediate the release of synaptic vesicles containing neurotransmitters in neurons. These neuronal SNAREs are the targets of the neurotoxins responsible for botulism and tetanus produced by certain bacteria.

Poneratoxin is a paralyzing neurotoxic peptide made by the bullet ant Paraponera clavata. It prevents inactivation of voltage gated sodium channels and therefore blocks synaptic transmission in the central nervous system. Specifically, poneratoxin acts on voltage gated sodium channels in skeletal muscle fibers, causing paralysis, and nociceptive fibers, causing pain. It is rated as a 4 plus on the Schmidt sting pain index, the highest possible rating with that system, and its effects can cause waves of pain up to twelve hours after a single sting. It is additionally being studied for its uses in biological insecticides.
A polyproline helix is a type of protein secondary structure which occurs in proteins comprising repeating proline residues. A left-handed polyproline II helix is formed when sequential residues all adopt (φ,ψ) backbone dihedral angles of roughly and have trans isomers of their peptide bonds. This PPII conformation is also common in proteins and polypeptides with other amino acids apart from proline. Similarly, a more compact right-handed polyproline I helix is formed when sequential residues all adopt (φ,ψ) backbone dihedral angles of roughly and have cis isomers of their peptide bonds. Of the twenty common naturally occurring amino acids, only proline is likely to adopt the cis isomer of the peptide bond, specifically the X-Pro peptide bond; steric and electronic factors heavily favor the trans isomer in most other peptide bonds. However, peptide bonds that replace proline with another N-substituted amino acid are also likely to adopt the cis isomer.

Voltage-gated potassium channels (VGKCs) are transmembrane channels specific for potassium and sensitive to voltage changes in the cell's membrane potential. During action potentials, they play a crucial role in returning the depolarized cell to a resting state.
Implicit solvation is a method to represent solvent as a continuous medium instead of individual “explicit” solvent molecules, most often used in molecular dynamics simulations and in other applications of molecular mechanics. The method is often applied to estimate free energy of solute-solvent interactions in structural and chemical processes, such as folding or conformational transitions of proteins, DNA, RNA, and polysaccharides, association of biological macromolecules with ligands, or transport of drugs across biological membranes.
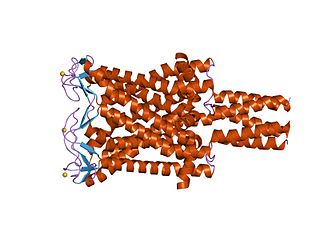
Large conductance mechanosensitive ion channels (MscLs) (TC# 1.A.22) are a family of pore-forming membrane proteins that are responsible for translating stresses at the cell membrane into an electrophysiological response. MscL has a relatively large conductance, 3 nS, making it permeable to ions, water, and small proteins when opened. MscL acts as stretch-activated osmotic release valve in response to osmotic shock.
Protein–lipid interaction is the influence of membrane proteins on the lipid physical state or vice versa.
Orientations of Proteins in Membranes (OPM) database provides spatial positions of membrane protein structures with respect to the lipid bilayer. Positions of the proteins are calculated using an implicit solvation model of the lipid bilayer. The results of calculations were verified against experimental studies of spatial arrangement of transmembrane and peripheral proteins in membranes.
Hydrophobicity scales are values that define the relative hydrophobicity or hydrophilicity of amino acid residues. The more positive the value, the more hydrophobic are the amino acids located in that region of the protein. These scales are commonly used to predict the transmembrane alpha-helices of membrane proteins. When consecutively measuring amino acids of a protein, changes in value indicate attraction of specific protein regions towards the hydrophobic region inside lipid bilayer.
Hydrophobic mismatch is the difference between the thicknesses of hydrophobic regions of a transmembrane protein and of the biological membrane it spans. In order to avoid unfavorable exposure of hydrophobic surfaces to water, the hydrophobic regions of transmembrane proteins are expected to have approximately the same thickness as the hydrophobic region of the surrounding lipid bilayer. Nevertheless, the same membrane protein can be encountered in bilayers of different thickness. In eukaryotic cells, the plasma membrane is thicker than the membranes of the endoplasmic reticulum. Yet all proteins that are abundant in the plasma membrane are initially integrated into the endoplasmic reticulum upon synthesis on ribosomes. Transmembrane peptides or proteins and surrounding lipids can adapt to the hydrophobic mismatch by different means.
Stephen H. White is an American Biophysicist, academic, and author. He is a Professor Emeritus of Physiology and Biophysics at the University of California, Irvine.

AmphipathicLipid Packing Sensor (ALPS) motifs were first identified in 2005 in ARFGAP1 and have been reviewed.

A single-pass membrane protein also known as single-spanning protein or bitopic protein is a transmembrane protein that spans the lipid bilayer only once. These proteins may constitute up to 50% of all transmembrane proteins, depending on the organism, and contribute significantly to the network of interactions between different proteins in cells, including interactions via transmembrane alpha helices. They usually include one or several water-soluble domains situated at the different sides of biological membranes, for example in single-pass transmembrane receptors. Some of them are small and serve as regulatory or structure-stabilizing subunits in large multi-protein transmembrane complexes, such as photosystems or the respiratory chain. A 2013 estimate identified about 1300 single-pass membrane proteins in the human genome.
Karen Renee Gibson Fleming is a Professor of Biophysics at Johns Hopkins University. She investigates the energetics of transmembrane helix-helix interactions. Fleming was awarded the 2020 Protein Society Carl Brändén Award.










