An inverted repeat is a single stranded sequence of nucleotides followed downstream by its reverse complement. The intervening sequence of nucleotides between the initial sequence and the reverse complement can be any length including zero. When the intervening length is zero, the composite sequence is a palindromic sequence. For example, 5'---TTACGnnnnnnCGTAA---3' is an inverted repeat sequence.

In molecular genetics, the three prime untranslated region (3'-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3'-UTR often contains regulatory regions that post-transcriptionally influence gene expression.

In genetics, a silencer is a DNA sequence capable of binding transcription regulation factors, called repressors. DNA contains genes and provides the template to produce messenger RNA (mRNA). That mRNA is then translated into proteins. When a repressor protein binds to the silencer region of DNA, RNA polymerase is prevented from transcribing the DNA sequence into RNA. With transcription blocked, the translation of RNA into proteins is impossible. Thus, silencers prevent genes from being expressed as proteins.
Long terminal repeats (LTRs) are identical sequences of DNA that repeat hundreds or thousands of times found at either end of retrotransposons or proviral DNA formed by reverse transcription of retroviral RNA. They are used by viruses to insert their genetic material into the host genomes.

This family represents the bovine leukaemia virus RNA encapsidation (packaging) signal, which is essential for efficient viral replication.

In molecular biology, the coronavirus frameshifting stimulation element is a conserved stem-loop of RNA found in coronaviruses that can promote ribosomal frameshifting. Such RNA molecules interact with a downstream region to form a pseudoknot structure; the region varies according to the virus but pseudoknot formation is known to stimulate frameshifting. In the classical situation, a sequence 32 nucleotides downstream of the stem is complementary to part of the loop. In other coronaviruses, however, another stem-loop structure around 150 nucleotides downstream can interact with members of this family to form kissing stem-loops and stimulate frameshifting.

The K10 transport/localisation element (TLS) is a 44 nucleotide K10 TLS regulatory element from Drosophila melanogaster. K10 TLS is responsible for the transport and anterior localisation of K10 mRNA and acts to establish dorsoventral polarity in the oocyte. It was discovered by Julia Serano.

The hairy localisation element (HLE) is an RNA element found in the 3' UTR of the hairy gene. HLE contains two stem-loops. HLE is essential for the mediation of apical localisation and the two stem-loop structures act to allow the recognition of hairy mRNA by the localisation machinery. HLE is found in Drosophila species.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.

Retroviral Psi packaging element is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.
The histone 3′ UTR stem-loop is an RNA element involved in nucleocytoplasmic transport of the histone mRNAs, and in the regulation of stability and of translation efficiency in the cytoplasm. The mRNAs of metazoan histone genes lack polyadenylation and a poly-A tail, instead 3′ end processing occurs at a site between this highly conserved stem-loop and a purine rich region around 20 nucleotides downstream. The stem-loop is bound by a 31 kDa stem-loop binding protein. Together with U7 snRNA binding of the HDE, SLBP binding nucleates the formation of the processing complex.
The HIV-1 Rev response element (RRE) is a highly structured, ~350 nucleotide RNA segment present in the Env coding region of unspliced and partially spliced viral mRNAs. In the presence of the HIV-1 accessory protein Rev, HIV-1 mRNAs that contain the RRE can be exported from the nucleus to the cytoplasm for downstream events such as translation and virion packaging.

Usually found in gram-positive bacteria, the T box leader sequence is an RNA element that controls gene expression through the regulation of translation by binding directly to a specific tRNA and sensing its aminoacylation state. This interaction controls expression of downstream aminoacyl-tRNA synthetase genes, amino acid biosynthesis, and uptake-related genes in a negative feedback loop. The uncharged tRNA acts as the effector for transcription antitermination of genes in the T-box leader family. The anticodon of a specific tRNA base pairs to a specifier sequence within the T-box motif, and the NCCA acceptor tail of the tRNA base pairs to a conserved bulge in the T-box antiterminator hairpin.

The Gurken localisation signal is an RNA regulatory element conserved across many species of the fruitfly genus Drosophila. The element consists of a RNA stem loop within the coding region that forms a signal for dynein-mediated Gurken mRNA transport to a cap near the oocyte nucleus.

The HBV RNA encapsidation signal epsilon is an element essential for HBV virus replication.
The Hepatitis B virus PRE stem-loop alpha is an RNA structure that is shown to play a role in nuclear export of HBV mRNAs.
The Hepatitis B virus PRE stem-loop beta is an RNA structure that is shown to play a role in nuclear export of HBV mRNAs.
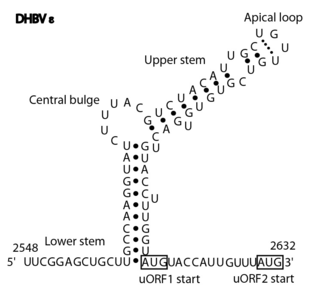
The Avian HBV RNA encapsidation signal epsilon is an RNA structure that is shown to facilitate encapsidation of the pregenomic RNA required for viral replication. There are two main classes of encapsidation signals in avian hepatitis B viruses - Duck hepatitis B virus (DHBV) and Heron hepatitis B virus (HHBV) like. DHBV is used as a model to understand human Hepatitis B virus. Although studies have shown that the HHBV epsilon has less pairing in the upper stem than DHBV, this pairing is not absolutely required for DHBV infection in ducks.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.














