
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.

A primer is a short single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. A synthetic primer may also be referred to as an oligo, short for oligonucleotide. DNA polymerase enzymes are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. DNA polymerase adds nucleotides after binding to the RNA primer and synthesizes the whole strand. Later, the RNA strands must be removed accurately and replace them with DNA nucleotides forming a gap region known as a nick that is filled in using an enzyme called ligase. The removal process of the RNA primer requires several enzymes, such as Fen1, Lig1, and others that work in coordination with DNA polymerase, to ensure the removal of the RNA nucleotides and the addition of DNA nucleotides. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and molecular biology that require in vitro DNA synthesis usually use DNA primers, since they are more temperature stable. Primers can be designed in laboratory for specific reactions such as polymerase chain reaction (PCR). When designing PCR primers, there are specific measures that must be taken into consideration, like the melting temperature of the primers and the annealing temperature of the reaction itself. Moreover, the DNA binding sequence of the primer in vitro has to be specifically chosen, which is done using a method called basic local alignment search tool (BLAST) that scans the DNA and finds specific and unique regions for the primer to bind.
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering.
In molecular biology, an amplicon is a piece of DNA or RNA that is the source and/or product of amplification or replication events. It can be formed artificially, using various methods including polymerase chain reactions (PCR) or ligase chain reactions (LCR), or naturally through gene duplication. In this context, amplification refers to the production of one or more copies of a genetic fragment or target sequence, specifically the amplicon. As it refers to the product of an amplification reaction, amplicon is used interchangeably with common laboratory terms, such as "PCR product."
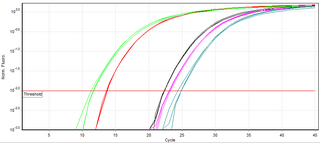
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.

Acrydite is a phosphoramidite that allows the synthesis of oligonucleotides with a methacryl group at the 5' end. Acryl oligonucleotides have been tested, but the acryl group is not stable to storage. Acrydite-modified oligonucleotides can react with nucleophiles such as thiols, this forms the basis of the ez-rays chemistry which was used for microarrays. More importantly, Acrydite-modified oligonucleotides can be incorporated, stoichiometrically, into hydrogels such as polyacrylamide, using standard free radical polymerization chemistry, where the double bond in the Acrydite group reacts with other activated double bond containing compounds such as acrylamide.

Cascade is an unincorporated community and U.S. Post Office in El Paso County, Colorado, United States. The ZIP Code of the Cascade Post Office is 80809.
TaqMan probes are hydrolysis probes that are designed to increase the specificity of quantitative PCR. The method was first reported in 1991 by researcher Kary Mullis at Cetus Corporation, and the technology was subsequently developed by Hoffmann-La Roche for diagnostic assays and by Applied Biosystems for research applications.
Nucleic acid thermodynamics is the study of how temperature affects the nucleic acid structure of double-stranded DNA (dsDNA). The melting temperature (Tm) is defined as the temperature at which half of the DNA strands are in the random coil or single-stranded (ssDNA) state. Tm depends on the length of the DNA molecule and its specific nucleotide sequence. DNA, when in a state where its two strands are dissociated, is referred to as having been denatured by the high temperature.
Multiplex ligation-dependent probe amplification (MLPA) is a variation of the multiplex polymerase chain reaction that permits amplification of multiple targets with only a single primer pair. It detects copy number changes at the molecular level, and software programs are used for analysis. Identification of deletions or duplications can indicate pathogenic mutations, thus MLPA is an important diagnostic tool used in clinical pathology laboratories worldwide.
The polymerase chain reaction (PCR) is a commonly used molecular biology tool for amplifying DNA, and various techniques for PCR optimization which have been developed by molecular biologists to improve PCR performance and minimize failure.
Loop-mediated isothermal amplification (LAMP) is a single-tube technique for the amplification of DNA and a low-cost alternative to detect certain diseases. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) combines LAMP with a reverse transcription step to allow the detection of RNA.

Eukaryotic translation initiation factor 4 gamma 1 is a protein that in humans is encoded by the EIF4G1 gene.

A nucleic acid test (NAT) is a technique used to detect a particular nucleic acid sequence and thus usually to detect and identify a particular species or subspecies of organism, often a virus or bacterium that acts as a pathogen in blood, tissue, urine, etc. NATs differ from other tests in that they detect genetic materials rather than antigens or antibodies. Detection of genetic materials allows an early diagnosis of a disease because the detection of antigens and/or antibodies requires time for them to start appearing in the bloodstream. Since the amount of a certain genetic material is usually very small, many NATs include a step that amplifies the genetic material—that is, makes many copies of it. Such NATs are called nucleic acid amplification tests (NAATs). There are several ways of amplification, including polymerase chain reaction (PCR), strand displacement assay (SDA), or transcription mediated assay (TMA).
The versatility of polymerase chain reaction (PCR) has led to modifications of the basic protocol being used in a large number of variant techniques designed for various purposes. This article summarizes many of the most common variations currently or formerly used in molecular biology laboratories; familiarity with the fundamental premise by which PCR works and corresponding terms and concepts is necessary for understanding these variant techniques.
Primer Premier software combines design of primers for various PCR applications under one common platform. The software supports design of degenerate primers on alignments for amplifying a related set of nucleotide sequences for detecting many common traits amongst organisms and to determine heredity.
OLIGO Primer Analysis Software is a software for DNA primer design. The first paper describing this software was published in 1989. The program is a real time PCR primer and probe search and analysis tool, in addition to siRNA and molecular beacon searches, open reading frame, restriction enzyme analysis. It was created and maintained by Wojciech Rychlik and Piotr Rychlik.
A primer dimer (PD) is a potential by-product in the polymerase chain reaction (PCR), a common biotechnological method. As its name implies, a PD consists of two primer molecules that have attached (hybridized) to each other because of strings of complementary bases in the primers. As a result, the DNA polymerase amplifies the PD, leading to competition for PCR reagents, thus potentially inhibiting amplification of the DNA sequence targeted for PCR amplification. In quantitative PCR, PDs may interfere with accurate quantification.
Multiplex polymerase chain reaction refers to the use of polymerase chain reaction to amplify several different DNA sequences simultaneously. This process amplifies DNA in samples using multiple primers and a temperature-mediated DNA polymerase in a thermal cycler. The primer design for all primers pairs has to be optimized so that all primer pairs can work at the same annealing temperature during PCR.

In silico PCR refers to computational tools used to calculate theoretical polymerase chain reaction (PCR) results using a given set of primers (probes) to amplify DNA sequences from a sequenced genome or transcriptome.







