
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

Structural biology is a field that is many centuries old which, as defined by the Journal of Structural Biology, deals with structural analysis of living material at every level of organization. Early structural biologists throughout the 19th and early 20th centuries were primarily only able to study structures to the limit of the naked eye's visual acuity and through magnifying glasses and light microscopes.

Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure. A number of tertiary structures may fold into a quaternary structure.
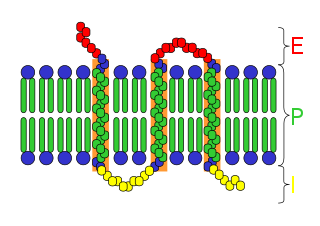
An integral, or intrinsic, membrane protein (IMP) is a type of membrane protein that is permanently attached to the biological membrane. All transmembrane proteins can be classified as IMPs, but not all IMPs are transmembrane proteins. IMPs comprise a significant fraction of the proteins encoded in an organism's genome. Proteins that cross the membrane are surrounded by annular lipids, which are defined as lipids that are in direct contact with a membrane protein. Such proteins can only be separated from the membranes by using detergents, nonpolar solvents, or sometimes denaturing agents.

Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid monomer may also be called a residue, which indicates a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions, such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, cryo-electron microscopy (cryo-EM) and dual polarisation interferometry, to determine the structure of proteins.

Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.

Photosystem II is the first protein complex in the light-dependent reactions of oxygenic photosynthesis. It is located in the thylakoid membrane of plants, algae, and cyanobacteria. Within the photosystem, enzymes capture photons of light to energize electrons that are then transferred through a variety of coenzymes and cofactors to reduce plastoquinone to plastoquinol. The energized electrons are replaced by oxidizing water to form hydrogen ions and molecular oxygen.

Robert Huber is a German biochemist and Nobel laureate. known for his work crystallizing an intramembrane protein important in photosynthesis and subsequently applying X-ray crystallography to elucidate the protein's structure.

Hartmut Michel is a German biochemist, who received the 1988 Nobel Prize in Chemistry for determination of the first crystal structure of an integral membrane protein, a membrane-bound complex of proteins and co-factors that is essential to photosynthesis.

Tetracycline-controlled transcriptional activation is a method of inducible gene expression where transcription is reversibly turned on or off in the presence of the antibiotic tetracycline or one of its derivatives.
The history of molecular biology begins in the 1930s with the convergence of various, previously distinct biological and physical disciplines: biochemistry, genetics, microbiology, virology and physics. With the hope of understanding life at its most fundamental level, numerous physicists and chemists also took an interest in what would become molecular biology.

Tet Repressor proteins are proteins playing an important role in conferring antibiotic resistance to large categories of bacterial species.

Biomolecular structure is the intricate folded, three-dimensional shape that is formed by a molecule of protein, DNA, or RNA, and that is important to its function. The structure of these molecules may be considered at any of several length scales ranging from the level of individual atoms to the relationships among entire protein subunits. This useful distinction among scales is often expressed as a decomposition of molecular structure into four levels: primary, secondary, tertiary, and quaternary. The scaffold for this multiscale organization of the molecule arises at the secondary level, where the fundamental structural elements are the molecule's various hydrogen bonds. This leads to several recognizable domains of protein structure and nucleic acid structure, including such secondary-structure features as alpha helixes and beta sheets for proteins, and hairpin loops, bulges, and internal loops for nucleic acids. The terms primary, secondary, tertiary, and quaternary structure were introduced by Kaj Ulrik Linderstrøm-Lang in his 1951 Lane Medical Lectures at Stanford University.

Photosynthetic reaction centre proteins are main protein components of photosynthetic reaction centres (RCs) of bacteria and plants. They are transmembrane proteins embedded in the chloroplast thylakoid or bacterial cell membrane.

Helen Miriam Berman is a Board of Governors Professor of Chemistry and Chemical Biology at Rutgers University and a former director of the RCSB Protein Data Bank. A structural biologist, her work includes structural analysis of protein-nucleic acid complexes, and the role of water in molecular interactions. She is also the founder and director of the Nucleic Acid Database, and led the Protein Structure Initiative Structural Genomics Knowledgebase.
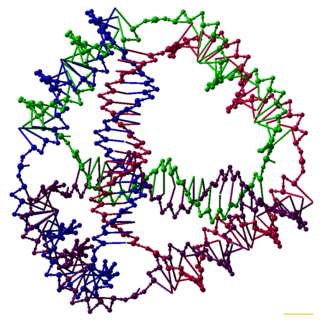
DNA nanotechnology is the design and manufacture of artificial nucleic acid structures for technological uses. In this field, nucleic acids are used as non-biological engineering materials for nanotechnology rather than as the carriers of genetic information in living cells. Researchers in the field have created static structures such as two- and three-dimensional crystal lattices, nanotubes, polyhedra, and arbitrary shapes, and functional devices such as molecular machines and DNA computers. The field is beginning to be used as a tool to solve basic science problems in structural biology and biophysics, including applications in X-ray crystallography and nuclear magnetic resonance spectroscopy of proteins to determine structures. Potential applications in molecular scale electronics and nanomedicine are also being investigated.

Macromolecular structure validation is the process of evaluating reliability for 3-dimensional atomic models of large biological molecules such as proteins and nucleic acids. These models, which provide 3D coordinates for each atom in the molecule, come from structural biology experiments such as x-ray crystallography or nuclear magnetic resonance (NMR). The validation has three aspects: 1) checking on the validity of the thousands to millions of measurements in the experiment; 2) checking how consistent the atomic model is with those experimental data; and 3) checking consistency of the model with known physical and chemical properties.
This is a timeline of crystallography.

Petra Fromme is a German-American chemist who is Director of the Biodesign Center for Applied Structural Discovery and Regents Professor at the Arizona State University. Her research considers the structure-to-function relationship of the membrane proteins involved with infectious diseases and bio-energy conversion. In 2021, she was awarded the Protein Society Anfinsen Award.














