Related Research Articles

In biology, histones are highly basic proteins found in eukaryotic cell nuclei that pack and order the DNA into structural units called nucleosomes. Histones are abundant in lysine and arginine. Histones are the chief protein components of chromatin, acting as spools around which DNA winds, and playing a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long. For example, each human diploid cell has about 1.8 meters of DNA; wound on the histones, the diploid cell has about 90 micrometers (0.09 mm) of chromatin. When the diploid cells are duplicated and condensed during mitosis, the result is about 120 micrometers of chromosomes.

Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.
Acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl functional group into a chemical compound. Such compounds are termed acetate esters or acetates. Deacetylation is the opposite reaction, the removal of an acetyl group from a chemical compound.
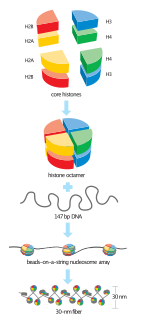
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes.
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes.

Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminal tail protruding from the histone core of the nucleosome are acetylated and deacetylated as part of gene regulation.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

Histone deacetylase 3 is an enzyme encoded by the HDAC3 gene in both humans and mice.

Peregrin also known as bromodomain and PHD finger-containing protein 1 is a protein that in humans is encoded by the BRPF1 gene located on 3p26-p25. Peregrin is a multivalent chromatin regulator that recognizes different epigenetic marks and activates three histone acetyltransferases. BRPF1 contains two PHD fingers, one bromodomain and one chromo/Tudor-related Pro-Trp-Trp-Pro (PWWP) domain.
H3K27ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 27th lysine residue of the histone H3 protein.
H2BK5ac is an epigenetic modification to the DNA packaging protein Histone H2B. It is a mark that indicates the acetylation at the 5th lysine residue of the histone H2B protein. H2BK5ac is involved in maintaining stem cells and colon cancer.
H4K16ac is an epigenetic modification to the DNA packaging protein Histone H4. It is a mark that indicates the acetylation at the 16th lysine residue of the histone H4 protein.
H4K5ac is an epigenetic modification to the DNA packaging protein histone H4. It is a mark that indicates the acetylation at the 5th lysine residue of the histone H4 protein. H4K5 is the closest lysine residue to the N-terminal tail of histone H4. It is enriched at the transcription start site (TSS) and along gene bodies. Acetylation of histone H4K5 and H4K12ac is enriched at centromeres.
H4K8ac, representing an epigenetic modification to the DNA packaging protein histone H4, is a mark indicating the acetylation at the 8th lysine residue of the histone H4 protein. It has been implicated in the prevalence of malaria.
H4K12ac is an epigenetic modification to the DNA packaging protein histone H4. It is a mark that indicates the acetylation at the 12th lysine residue of the histone H4 protein. H4K12ac is involved in learning and memory. It is possible that restoring this modification could reduce age-related decline in memory.
H4K91ac is an epigenetic modification to the DNA packaging protein histone H4. It is a mark that indicates the acetylation at the 91st lysine residue of the histone H4 protein. No known diseases are attributed to this mark but it might be implicated in melanoma.
H3K23ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 23rd lysine residue of the histone H3 protein.
H3K14ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 14th lysine residue of the histone H3 protein.
H3K9ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 9th lysine residue of the histone H3 protein.
H3K56ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 56th lysine residue of the histone H3 protein.
References
- ↑ Schulze JM, Wang AY, Kobor MS (2009). "YEATS domain proteins: a diverse family with many links to chromatin modification and transcription". Biochem. Cell Biol. 87 (1): 65–75. doi:10.1139/O08-111. PMID 19234524.
- ↑ Wang AY, Schulze JM, Skordalakes E, Gin JW, Berger JM, Rine J, Kobor MS (2009). "Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation". Proc. Natl. Acad. Sci. U.S.A. 106 (51): 21573–8. doi:10.1073/pnas.0906539106. PMC 2799888 . PMID 19966225.
- ↑ Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SY, Li W, Li H, Shi X (2014). "AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation". Cell. 159 (3): 558–71. doi:10.1016/j.cell.2014.09.049. PMC 4344132 . PMID 25417107.
- ↑ Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, Shi X, Strahl BD, Kutateladze TG (2016). "The Taf14 YEATS domain is a reader of histone crotonylation". Nat. Chem. Biol. 12 (6): 396–8. doi:10.1038/nchembio.2065. PMC 4871749 . PMID 27089029.