
Pseudomonadota is a major phylum of Gram-negative bacteria. The renaming of several prokaryote phyla in 2021, including Pseudomonadota, remains controversial among microbiologists, many of whom continue to use the earlier name Proteobacteria, of long standing in the literature. The phylum Proteobacteria includes a wide variety of pathogenic genera, such as Escherichia, Salmonella, Vibrio, Yersinia, Legionella, and many others. Others are free-living (non-parasitic) and include many of the bacteria responsible for nitrogen fixation.

Rickettsia is a genus of nonmotile, gram-negative, nonspore-forming, highly pleomorphic bacteria that may occur in the forms of cocci, bacilli, or threads. The genus was named after Howard Taylor Ricketts in honor of his pioneering work on tick-borne spotted fever.

The Campylobacterales are an order of Campylobacterota which make up the epsilon subdivision, together with the small family Nautiliaceae. They are Gram-negative. Most of the species are microaerophilic.

The Rickettsiales, informally called rickettsias, are an order of small Alphaproteobacteria. They are obligate intracellular parasites, and some are notable pathogens, including Rickettsia, which causes a variety of diseases in humans, and Ehrlichia, which causes diseases in livestock. Another genus of well-known Rickettsiales is the Wolbachia, which infect about two-thirds of all arthropods and nearly all filarial nematodes. Genetic studies support the endosymbiotic theory according to which mitochondria and related organelles developed from members of this group.

Caulobacteraceae is a family of Pseudomonadota within the alpha subgroup. Like all Pseudomonadota, the Caulobacteraceae are gram-negative. Caulobacteraceae includes the genera Asticcacaulis, Brevundimonas, Phenylobacterium and Caulobacter.

The Hyphomicrobiales are an order of Gram-negative Alphaproteobacteria.

The Rhodocyclales are an order of the class Betaproteobacteria in the phylum "Pseudomonadota". Following a major reclassification of the class in 2017, the previously monofamilial order was split into three families:

The Comamonadaceae are a family of the Betaproteobacteria. Like all Pseudomonadota, they are Gram-negative. They are aerobic and most of the species are motile via flagella. The cells are curved rod-shaped.

The Rickettsiaceae are a family of bacteria. The genus Rickettsia is the most prominent genus within the family. The bacteria that eventually formed the mitochondrion is believed to have originated from this family. Most human pathogens in this family are in genus Rickettsia. They spend part of their lifecycle in the bodies of arthropods such as ticks or lice, and are then transmitted to humans or other mammals by the bite of the arthropod. It contains Gram-negative bacteria, very sensitive to environmental exposure, thus is adapted to obligate intracellular infection. Rickettsia rickettsii is considered the prototypical infectious organism in the group.

Lassa mammarenavirus (LASV) is an arenavirus that causes Lassa hemorrhagic fever, a type of viral hemorrhagic fever (VHF), in humans and other primates. Lassa mammarenavirus is an emerging virus and a select agent, requiring Biosafety Level 4-equivalent containment. It is endemic in West African countries, especially Sierra Leone, the Republic of Guinea, Nigeria, and Liberia, where the annual incidence of infection is between 300,000 and 500,000 cases, resulting in 5,000 deaths per year.

Betaproteobacteria are a class of Gram-negative bacteria, and one of the eight classes of the phylum Pseudomonadota.

Alphaproteobacteria is a class of bacteria in the phylum Pseudomonadota. The Magnetococcales and Mariprofundales are considered basal or sister to the Alphaproteobacteria. The Alphaproteobacteria are highly diverse and possess few commonalities, but nevertheless share a common ancestor. Like all Proteobacteria, its members are gram-negative and some of its intracellular parasitic members lack peptidoglycan and are consequently gram variable.

The SAM-II riboswitch is an RNA element found predominantly in Alphaproteobacteria that binds S-adenosyl methionine (SAM). Its structure and sequence appear to be unrelated to the SAM riboswitch found in Gram-positive bacteria. This SAM riboswitch is located upstream of the metA and metC genes in Agrobacterium tumefaciens, and other methionine and SAM biosynthesis genes in other alpha-proteobacteria. Like the other SAM riboswitch, it probably functions to turn off expression of these genes in response to elevated SAM levels. A significant variant of SAM-II riboswitches was found in Pelagibacter ubique and related marine bacteria and called SAM-V. Also, like many structured RNAs, SAM-II riboswitches can tolerate long loops between their stems.

SerC leader is a putative regulatory RNA structure found upstream of the serC-serA operon in some alpha-proteobacteria. The final stem of the structure overlaps the ribosome binding site of the serC reading frame.
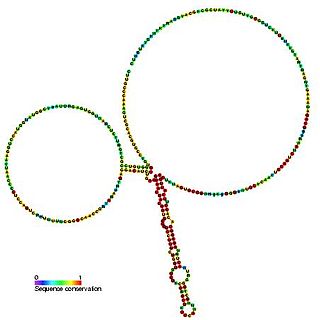
SpeF is a putative cis-acting element identified in several gram negative alpha proteobacteria. It is proposed to be involved in regulating expression of genes involved in polyamide biosynthesis.

suhB, also known as mmgR, is a non-coding RNA found multiple times in the Agrobacterium tumefaciens genome and related alpha-proteobacteria. Other non-coding RNAs uncovered in the same analysis include speF, ybhL, metA, and serC.
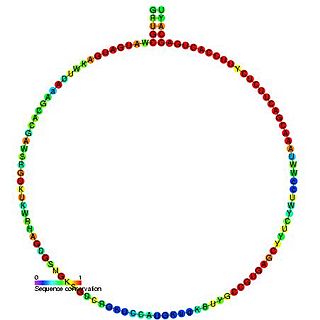
In molecular biology, Small nucleolar RNA Z152 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z152 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA Z152 was identified in screens of Oryza sativa and Arabidopsis thaliana.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.
sRNA-Xcc1 is a family of trans-acting non-coding RNA. Homologs of sRNA-Xcc1 are found in a few bacterial strains belonging to alpha-proteobacteria, beta-proteobacteria, gamma-proteobacteria, and delta-proteobacteria. In Xanthomonascampestris pv. campestris, sRNA-Xcc1 is encoded by an integron gene cassette and is under the positive control of the virulence regulators HrpG and HrpX.
















