GTPases are a large family of hydrolase enzymes that bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). The GTP binding and hydrolysis takes place in the highly conserved P-loop "G domain", a protein domain common to many GTPases.
The signal recognition particle (SRP) is an abundant, cytosolic, universally conserved ribonucleoprotein that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and the plasma membrane in prokaryotes.
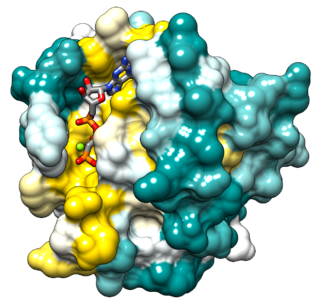
Ras, from "Rat sarcoma virus", is a family of related proteins that are expressed in all animal cell lineages and organs. All Ras protein family members belong to a class of protein called small GTPase, and are involved in transmitting signals within cells. Ras is the prototypical member of the Ras superfamily of proteins, which are all related in three-dimensional structure and regulate diverse cell behaviours.
Small GTPases, also known as small G-proteins, are a family of hydrolase enzymes that can bind and hydrolyze guanosine triphosphate (GTP). They are a type of G-protein found in the cytosol that are homologous to the alpha subunit of heterotrimeric G-proteins, but unlike the alpha subunit of G proteins, a small GTPase can function independently as a hydrolase enzyme to bind to and hydrolyze a guanosine triphosphate (GTP) to form guanosine diphosphate (GDP). The best-known members are the Ras GTPases and hence they are sometimes called Ras subfamily GTPases.

ADP ribosylation factors (ARFs) are members of the ARF family of GTP-binding proteins of the Ras superfamily. ARF family proteins are ubiquitous in eukaryotic cells, and six highly conserved members of the family have been identified in mammalian cells. Although ARFs are soluble, they generally associate with membranes because of N-terminus myristoylation. They function as regulators of vesicular traffic and actin remodelling.
A release factor is a protein that allows for the termination of translation by recognizing the termination codon or stop codon in an mRNA sequence. They are named so because they release new peptides from the ribosome.

EF-Tu is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM.

Guanine nucleotide exchange factors (GEFs) are proteins or protein domains that activate monomeric GTPases by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP). A variety of unrelated structural domains have been shown to exhibit guanine nucleotide exchange activity. Some GEFs can activate multiple GTPases while others are specific to a single GTPase.
Eukaryotic initiation factors (eIFs) are proteins or protein complexes involved in the initiation phase of eukaryotic translation. These proteins help stabilize the formation of ribosomal preinitiation complexes around the start codon and are an important input for post-transcription gene regulation. Several initiation factors form a complex with the small 40S ribosomal subunit and Met-tRNAiMet called the 43S preinitiation complex. Additional factors of the eIF4F complex recruit the 43S PIC to the five-prime cap structure of the mRNA, from which the 43S particle scans 5'-->3' along the mRNA to reach an AUG start codon. Recognition of the start codon by the Met-tRNAiMet promotes gated phosphate and eIF1 release to form the 48S preinitiation complex, followed by large 60S ribosomal subunit recruitment to form the 80S ribosome. There exist many more eukaryotic initiation factors than prokaryotic initiation factors, reflecting the greater biological complexity of eukaryotic translation. There are at least twelve eukaryotic initiation factors, composed of many more polypeptides, and these are described below.
Elongation factor 4 (EF-4) is an elongation factor that is thought to back-translocate on the ribosome during the translation of RNA to proteins. It is found near-universally in bacteria and in eukaryotic endosymbiotic organelles including the mitochondria and the plastid. Responsible for proofreading during protein synthesis, EF-4 is a recent addition to the nomenclature of bacterial elongation factors.
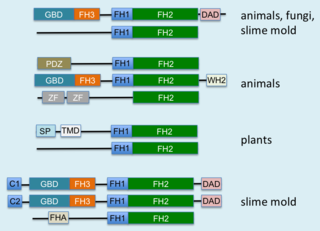
Formins (formin homology proteins) are a group of proteins that are involved in the polymerization of actin and associate with the fast-growing end (barbed end) of actin filaments. Most formins are Rho-GTPase effector proteins. Formins regulate the actin and microtubule cytoskeleton and are involved in various cellular functions such as cell polarity, cytokinesis, cell migration and SRF transcriptional activity. Formins are multidomain proteins that interact with diverse signalling molecules and cytoskeletal proteins, although some formins have been assigned functions within the nucleus.

Transforming protein RhoA, also known as Ras homolog family member A (RhoA), is a small GTPase protein in the Rho family of GTPases that in humans is encoded by the RHOA gene. While the effects of RhoA activity are not all well known, it is primarily associated with cytoskeleton regulation, mostly actin stress fibers formation and actomyosin contractility. It acts upon several effectors. Among them, ROCK1 and DIAPH1 are the best described. RhoA, and the other Rho GTPases, are part of a larger family of related proteins known as the Ras superfamily, a family of proteins involved in the regulation and timing of cell division. RhoA is one of the oldest Rho GTPases, with homologues present in the genomes since 1.5 billion years. As a consequence, RhoA is somehow involved in many cellular processes which emerged throughout evolution. RhoA specifically is regarded as a prominent regulatory factor in other functions such as the regulation of cytoskeletal dynamics, transcription, cell cycle progression and cell transformation.

Ras-related protein Rap-1A is a protein that in humans is encoded by the RAP1A gene.

RAN binding protein 2 (RANBP2) is protein which in humans is encoded by the RANBP2 gene. It is also known as nucleoporin 358 (Nup358) since it is a member nucleoporin family that makes up the nuclear pore complex. RanBP2 has a mass of 358 kDa.
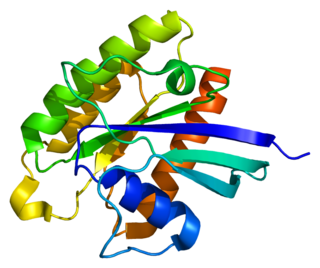
RHEB also known as Ras homolog enriched in brain (RHEB) is a GTP-binding protein that is ubiquitously expressed in humans and other mammals. The protein is largely involved in the mTOR pathway and the regulation of the cell cycle.
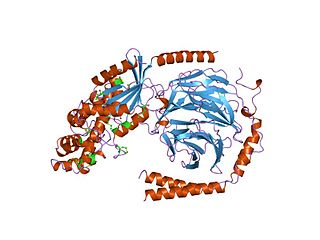
G alpha subunits are one of the three types of subunit of guanine nucleotide binding proteins, which are membrane-associated, heterotrimeric G proteins.

RhoG is a small monomeric GTP-binding protein, and is an important component of many intracellular signalling pathways. It is a member of the Rac subfamily of the Rho family of small G proteins and is encoded by the gene RHOG.

EF-G is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.
EF-Ts is one of the prokaryotic elongation factors. It is found in human mitochondria as TSFM. It is similar to eukaryotic EF-1B.
Alfred "Fred" Wittinghofer is a German biochemist.












