Related Research Articles

Drug resistance is the reduction in effectiveness of a medication such as an antimicrobial or an antineoplastic in treating a disease or condition. The term is used in the context of resistance that pathogens or cancers have "acquired", that is, resistance has evolved. Antimicrobial resistance and antineoplastic resistance challenge clinical care and drive research. When an organism is resistant to more than one drug, it is said to be multidrug-resistant.

The ATP-binding cassette transporters are a transport system superfamily that is one of the largest and possibly one of the oldest gene families. It is represented in all extant phyla, from prokaryotes to humans.

P-glycoprotein 1 also known as multidrug resistance protein 1 (MDR1) or ATP-binding cassette sub-family B member 1 (ABCB1) or cluster of differentiation 243 (CD243) is an important protein of the cell membrane that pumps many foreign substances out of cells. More formally, it is an ATP-dependent efflux pump with broad substrate specificity. It exists in animals, fungi, and bacteria, and it likely evolved as a defense mechanism against harmful substances.
Glycylcyclines are a class of antibiotics derived from tetracycline. These tetracycline analogues are specifically designed to overcome two common mechanisms of tetracycline resistance, namely resistance mediated by acquired efflux pumps and/or ribosomal protection. Presently, tigecycline is the only glycylcycline approved for antibiotic use.

Lincosamides are a class of antibiotics, which include lincomycin, clindamycin, and pirlimycin.
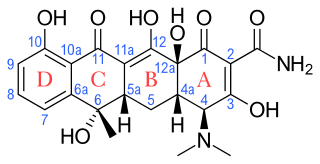
Tetracyclines are a group of broad-spectrum antibiotic compounds that have a common basic structure and are either isolated directly from several species of Streptomyces bacteria or produced semi-synthetically from those isolated compounds. Tetracycline molecules comprise a linear fused tetracyclic nucleus to which a variety of functional groups are attached. Tetracyclines are named for their four ("tetra-") hydrocarbon rings ("-cycl-") derivation ("-ine"). They are defined as a subclass of polyketides, having an octahydrotetracene-2-carboxamide skeleton and are known as derivatives of polycyclic naphthacene carboxamide. While all tetracyclines have a common structure, they differ from each other by the presence of chloride, methyl, and hydroxyl groups. These modifications do not change their broad antibacterial activity, but do affect pharmacological properties such as half-life and binding to proteins in serum.
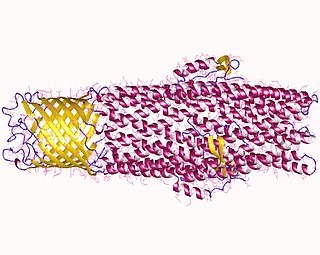
All microorganisms, with a few exceptions, have highly conserved DNA sequences in their genome that are transcribed and translated to efflux pumps. Efflux pumps are capable of moving a variety of different toxic compounds out of cells, such as antibiotics, heavy metals, organic pollutants, plant-produced compounds, quorum sensing signals, bacterial metabolites and neurotransmitters via active efflux, which is vital part for xenobiotic metabolism. This active efflux mechanism is responsible for various types of resistance to bacterial pathogens within bacterial species - the most concerning being antibiotic resistance because microorganisms can have adapted efflux pumps to divert toxins out of the cytoplasm and into extracellular media.
Cross-resistance is when something develops resistance to several substances that have a similar mechanism of action. For example, say a certain type of bacteria develops resistance to one antibiotic. That bacteria then also has resistance to several other antibiotics that target the same protein or use the same route to get into the bacterium. A real example of cross-resistance occurred for nalidixic acid and ciprofloxacin, which are both quinolone antibiotics. When bacteria developed resistance to ciprofloxacin, they also developed resistance to nalidixic acid because both drugs work by inhibiting topoisomerase, a key enzyme in DNA replication. Due to cross-resistance, antimicrobial treatments like phage therapy can quickly lose their efficacy against bacteria.

ATP-binding cassette transporter sub-family C member 11, also MRP8 is a membrane transporter that exports certain molecules from inside a cell. It is a protein that in humans is encoded by gene ABCC11.

Multidrug and toxin extrusion protein 2 is a protein which in humans is encoded by the SLC47A2 gene.
Multi-antimicrobial extrusion protein (MATE) also known as multidrug and toxin extrusion or multidrug and toxic compound extrusion is a family of proteins which function as drug/sodium or proton antiporters.

Neisseria gonorrhoeae, the bacterium that causes the sexually transmitted infection gonorrhea, has developed antibiotic resistance to many antibiotics. The bacteria was first identified in 1879.
The p-aminobenzoyl-glutamate transporter(AbgT) family is a family of transporter proteins belonging to the ion transporter (IT) superfamily. The AbgT family consists of the AbgT protein of E. coli and the MtrF drug exporter of Neisseria gonorrhoeae. The former protein is apparently cryptic in wild-type cells, but when expressed on a high copy number plasmid, or when expressed at higher levels due to mutation, it appeared to allow uptake and subsequent utilization of p-aminobenzoyl-glutamate as a source of p-aminobenzoate for p-aminobenzoate auxotrophs. p-Aminobenzoate is a constituent of and a precursor for the biosynthesis of folic acid. MtrF was annotated as a putative drug efflux pump.
Arsenite resistance (Ars) efflux pumps of bacteria may consist of two proteins, ArsB and ArsA, or of one protein. ArsA proteins have two ATP binding domains and probably arose by a tandem gene duplication event. ArsB proteins all possess twelve transmembrane spanners and may also have arisen by a tandem gene duplication event. Structurally, the Ars pumps resemble ABC-type efflux pumps, but there is no significant sequence similarity between the Ars and ABC pumps. When only ArsB is present, the system operates by a pmf-dependent mechanism, and consequently belongs in TC subclass 2.A. When ArsA is also present, ATP hydrolysis drives efflux, and consequently the system belongs in TC subclass 3.A. ArsB therefore appears twice in the TC system but ArsA appears only once. These pumps actively expel both arsenite and antimonite.
Multidrug resistance pumps also known Multidrug efflux pumps are a type of efflux pump and P-glycoprotein. MDR pumps in the cell membrane extrudes many foreign substances out of the cells and some pumps can have a broad specificity. MDR pumps exist in animals, fungi, and bacteria and likely evolved as a defense mechanism against harmful substances. There are seven families of MDRs and are grouped by homology, energy source, and overall structure.

Resistance-nodulation-division (RND) family transporters are a category of bacterial efflux pumps, especially identified in Gram-negative bacteria and located in the cytoplasmic membrane, that actively transport substrates. The RND superfamily includes seven families: the heavy metal efflux (HME), the hydrophobe/amphiphile efflux-1, the nodulation factor exporter family (NFE), the SecDF protein-secretion accessory protein family, the hydrophobe/amphiphile efflux-2 family, the eukaryotic sterol homeostasis family, and the hydrophobe/amphiphile efflux-3 family. These RND systems are involved in maintaining homeostasis of the cell, removal of toxic compounds, and export of virulence determinants. They have a broad substrate spectrum and can lead to the diminished activity of unrelated drug classes if over-expressed. The first reports of drug resistant bacterial infections were reported in the 1940s after the first mass production of antibiotics. Most of the RND superfamily transport systems are made of large polypeptide chains. RND proteins exist primarily in gram-negative bacteria but can also be found in gram-positive bacteria, archaea, and eukaryotes.
ESKAPE is an acronym comprising the scientific names of six highly virulent and antibiotic resistant bacterial pathogens including: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. This group of Gram-positive and Gram-negative bacteria can evade or 'escape' commonly used antibiotics due to their increasing multi-drug resistance (MDR). As a result, throughout the world, they are the major cause of life-threatening nosocomial or hospital-acquired infections in immunocompromised and critically ill patients who are most at risk. P. aeruginosa and S. aureus are some of the most ubiquitous pathogens in biofilms found in healthcare. P. aeruginosa is a Gram-negative, rod-shaped bacterium, commonly found in the gut flora, soil, and water that can be spread directly or indirectly to patients in healthcare settings. The pathogen can also be spread in other locations through contamination, including surfaces, equipment, and hands. The opportunistic pathogen can cause hospitalized patients to have infections in the lungs, blood, urinary tract, and in other body regions after surgery. S. aureus is a Gram-positive, cocci-shaped bacterium, residing in the environment and on the skin and nose of many healthy individuals. The bacterium can cause skin and bone infections, pneumonia, and other types of potentially serious infections if it enters the body. S. aureus has also gained resistance to many antibiotic treatments, making healing difficult. Because of natural and unnatural selective pressures and factors, antibiotic resistance in bacteria usually emerges through genetic mutation or acquires antibiotic-resistant genes (ARGs) through horizontal gene transfer - a genetic exchange process by which antibiotic resistance can spread.
Christine A. Hrycyna is a Professor of Biochemistry at Purdue University. She studies multi-drug resistance in human cancer, which usually occurs due to over expression of the MDR1 gene.

Multidrug-resistant bacteria are bacteria that are resistant to three or more classes of antimicrobial drugs. MDR bacteria have seen an increase in prevalence in recent years and pose serious risks to public health. MDR bacteria can be broken into 3 main categories: Gram-positive, Gram-negative, and other (acid-stain). These bacteria employ various adaptations to avoid or mitigate the damage done by antimicrobials. With increased access to modern medicine there has been a sharp increase in the amount of antibiotics consumed. Given the abundant use of antibiotics there has been a considerable increase in the evolution of antimicrobial resistance factors, now outpacing the development of new antibiotics.
Laura Piddock is a microbiologist, specialising in antibiotics and antibiotic resistance in bacteria. She is a Professor at the University of Birmingham, UK and also Scientific Director within the Global Antibiotic Research and Development Partnership.
References
- 1 2 Liu, B.; Pop, M. (2009-01-01). "ARDB--Antibiotic Resistance Genes Database". Nucleic Acids Research. 37 (Database): D443–D447. doi:10.1093/nar/gkn656. ISSN 0305-1048. PMC 2686595 . PMID 18832362.
- ↑ Higgins, Christopher F. (April 2007). "Multiple molecular mechanisms for multidrug resistance transporters". Nature. 446 (7137): 749–757. Bibcode:2007Natur.446..749H. doi:10.1038/nature05630. ISSN 0028-0836. PMID 17429392. S2CID 4414214.