
Ornithine transcarbamylase (OTC) is an enzyme that catalyzes the reaction between carbamoyl phosphate (CP) and ornithine (Orn) to form citrulline (Cit) and phosphate (Pi). There are two classes of OTC: anabolic and catabolic. This article focuses on anabolic OTC. Anabolic OTC facilitates the sixth step in the biosynthesis of the amino acid arginine in prokaryotes. In contrast, mammalian OTC plays an essential role in the urea cycle, the purpose of which is to capture toxic ammonia and transform it into urea, a less toxic nitrogen source, for excretion.

Glycine encephalopathy is a rare autosomal recessive disorder of glycine metabolism. After phenylketonuria, glycine encephalopathy is the second most common disorder of amino acid metabolism. The disease is caused by defects in the glycine cleavage system, an enzyme responsible for glycine catabolism. There are several forms of the disease, with varying severity of symptoms and time of onset. The symptoms are exclusively neurological in nature, and clinically this disorder is characterized by abnormally high levels of the amino acid glycine in bodily fluids and tissues, especially the cerebrospinal fluid.

Steroid 11β-hydroxylase, also known as steroid 11β-monooxygenase, is a steroid hydroxylase found in the zona glomerulosa and zona fasciculata of the adrenal cortex. Named officially the cytochrome P450 11B1, mitochondrial, it is a protein that in humans is encoded by the CYP11B1 gene. The enzyme is involved in the biosynthesis of adrenal corticosteroids by catalyzing the addition of hydroxyl groups during oxidation reactions.

Pyridoxine 5′-phosphate oxidase is an enzyme, encoded by the PNPO gene, that catalyzes several reactions in the vitamin B6 metabolism pathway. Pyridoxine 5′-phosphate oxidase catalyzes the final, rate-limiting step in vitamin B6 metabolism, the biosynthesis of pyridoxal 5′-phosphate, the biologically active form of vitamin B6 which acts as an essential cofactor. Pyridoxine 5′-phosphate oxidase is a member of the enzyme class oxidases, or more specifically, oxidoreductases. These enzymes catalyze a simultaneous oxidation-reduction reaction. The substrate oxidase enzymes is hydroxlyated by one oxygen atom of molecular oxygen. Concurrently, the other oxygen atom is reduced to water. Even though molecular oxygen is the electron acceptor in these enzymes' reactions, they are unique because oxygen does not appear in the oxidized product.
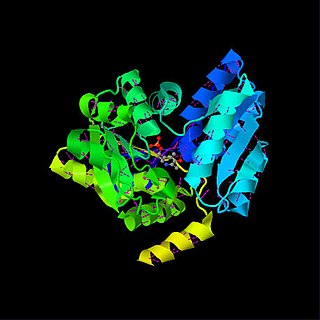
Serine dehydratase or L-serine ammonia lyase (SDH) is in the β-family of pyridoxal phosphate-dependent (PLP) enzymes. SDH is found widely in nature, but its structure and properties vary among species. SDH is found in yeast, bacteria, and the cytoplasm of mammalian hepatocytes. SDH catalyzes the deamination of L-serine to yield pyruvate, with the release of ammonia.

Glycine decarboxylase also known as glycine cleavage system P protein or glycine dehydrogenase is an enzyme that in humans is encoded by the GLDC gene.

The enzyme threonine aldolase is an enzyme that catalyzes the chemical reaction

Glucose-6-phosphatase, catalytic subunit is an enzyme that in humans is encoded by the G6PC gene.

Calcium-transporting ATPase type 2C member 1 is an enzyme that in humans is encoded by the ATP2C1 gene.

NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial is an enzyme that in humans is encoded by the NDUFS3 gene on chromosome 11. This gene encodes one of the iron-sulfur protein (IP) components of mitochondrial NADH:ubiquinone oxidoreductase. Mutations in this gene are associated with Leigh syndrome resulting from mitochondrial complex I deficiency.

NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial (NDUFS1) is an enzyme that in humans is encoded by the NDUFS1 gene. The encoded protein, NDUFS1, is the largest subunit of complex I, located on the inner mitochondrial membrane, and is important for mitochondrial oxidative phosphorylation. Mutations in this gene are associated with complex I deficiency.

NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial (NDUFV2) is an enzyme that in humans is encoded by the NDUFV2 gene. The encoded protein, NDUFV2, is a subunit of complex I of the mitochondrial respiratory chain, which is located on the inner mitochondrial membrane and involved in oxidative phosphorylation. Mutations in this gene are implicated in Parkinson's disease, bipolar disorder, schizophrenia, and have been found in one case of early onset hypertrophic cardiomyopathy and encephalopathy.

Glycine cleavage system H protein, mitochondrial is a protein that in humans is encoded by the GCSH gene. Degradation of glycine is brought about by the glycine cleavage system (GCS), which is composed of 4 protein components: P protein, H protein, T protein, and L protein. The H protein shuttles the methylamine group of glycine from the P protein to the T protein. The protein encoded by GCSH gene is the H protein, which transfers the methylamine group of glycine from the P protein to the T protein. Defects in this gene are a cause of nonketotic hyperglycinemia (NKH). Two transcript variants, one protein-coding and the other probably not protein-coding, have been found for this gene. Also, several transcribed and non-transcribed pseudogenes of this gene exist throughout the genome.

NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial is an enzyme that in humans is encoded by the NDUFS6 gene.
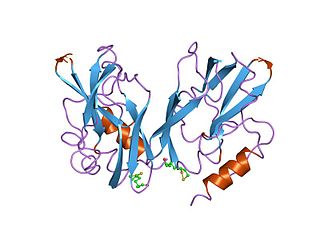
The glycine cleavage system (GCS) is also known as the glycine decarboxylase complex or GDC. The system is a series of enzymes that are triggered in response to high concentrations of the amino acid glycine. The same set of enzymes is sometimes referred to as glycine synthase when it runs in the reverse direction to form glycine. The glycine cleavage system is composed of four proteins: the T-protein, P-protein, L-protein, and H-protein. They do not form a stable complex, so it is more appropriate to call it a "system" instead of a "complex". The H-protein is responsible for interacting with the three other proteins and acts as a shuttle for some of the intermediate products in glycine decarboxylation. In both animals and plants, the glycine cleavage system is loosely attached to the inner membrane of the mitochondria. Mutations in this enzymatic system are linked with glycine encephalopathy.
Enolase Deficiency is a rare genetic disorder of glucose metabolism. Partial deficiencies have been observed in several caucasian families. The deficiency is transmitted through an autosomal dominant inheritance pattern. The gene for Enolase 1 has been localized to Chromosome 1 in humans. Enolase deficiency, like other glycolytic enzyme deficiences, usually manifests in red blood cells as they rely entirely on anaerobic glycolysis. Enolase deficiency is associated with a spherocytic phenotype and can result in hemolytic anemia, which is responsible for the clinical signs of Enolase deficiency.

Aldehyde dehydrogenase 7 family, member A1, also known as ALDH7A1 or antiquitin, is an enzyme that in humans is encoded by the ALDH7A1 gene. The protein encoded by this gene is a member of subfamily 7 in the aldehyde dehydrogenase gene family. These enzymes are thought to play a major role in the detoxification of aldehydes generated by alcohol metabolism and lipid peroxidation. This particular member has homology to a previously described protein from the green garden pea, the 26g pea turgor protein. It is also involved in lysine catabolism that is known to occur in the mitochondrial matrix. Recent reports show that this protein is found both in the cytosol and the mitochondria, and the two forms likely arise from the use of alternative translation initiation sites. An additional variant encoding a different isoform has also been found for this gene. Mutations in this gene are associated with pyridoxine-dependent epilepsy. Several related pseudogenes have also been identified.

Glutaredoxin 5, also known as GLRX5, is a protein which in humans is encoded by the GLRX5 gene located on chromosome 14. This gene encodes a mitochondrial protein, which is evolutionarily conserved. It is involved in the biogenesis of iron- sulfur clusters, which are required for normal iron homeostasis. Mutations in this gene are associated with autosomal recessive pyridoxine-refractory sideroblastic anemia.
Lipoate–protein ligase (EC 2.7.7.63, LplA, lipoate protein ligase, lipoate–protein ligase A, LPL, LPL-B) is an enzyme with systematic name ATP:lipoate adenylyltransferase. This enzyme catalyses the following chemical reaction

Lipoic acid synthetase is a protein that in humans is encoded by the LIAS gene.























