
Genetics is a branch of biology concerned with the study of genes, genetic variation, and heredity in organisms.

Genetic engineering, also called genetic modification or genetic manipulation, is the direct manipulation of an organism's genes using biotechnology. It is a set of technologies used to change the genetic makeup of cells, including the transfer of genes within and across species boundaries to produce improved or novel organisms. New DNA is obtained by either isolating and copying the genetic material of interest using recombinant DNA methods or by artificially synthesising the DNA. A construct is usually created and used to insert this DNA into the host organism. The first recombinant DNA molecule was made by Paul Berg in 1972 by combining DNA from the monkey virus SV40 with the lambda virus. As well as inserting genes, the process can be used to remove, or "knock out", genes. The new DNA can be inserted randomly, or targeted to a specific part of the genome.

In the fields of molecular biology and genetics, a genome is all genetic material of an organism. It consists of DNA. The genome includes both the genes and the noncoding DNA, as well as mitochondrial DNA and chloroplast DNA. The study of the genome is called genomics.

Reproduction is the biological process by which new individual organisms – "offspring" – are produced from their "parent" or parents. Reproduction is a fundamental feature of all known life; each individual organism exists as the result of reproduction. There are two forms of reproduction: asexual and sexual.
Non-coding DNA sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules. Other functions of non-coding DNA include the transcriptional and translational regulation of protein-coding sequences, scaffold attachment regions, origins of DNA replication, centromeres and telomeres. Its RNA counterpart is non-coding RNA.
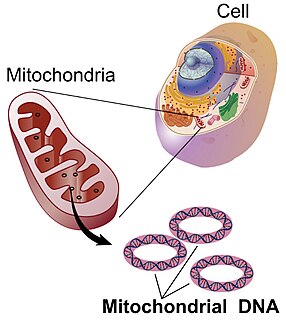
Mitochondrial DNA is the DNA located in mitochondria, cellular organelles within eukaryotic cells that convert chemical energy from food into a form that cells can use, adenosine triphosphate (ATP). Mitochondrial DNA is only a small portion of the DNA in a eukaryotic cell; most of the DNA can be found in the cell nucleus and, in plants and algae, also in plastids such as chloroplasts.

Genome projects are scientific endeavours that ultimately aim to determine the complete genome sequence of an organism and to annotate protein-coding genes and other important genome-encoded features. The genome sequence of an organism includes the collective DNA sequences of each chromosome in the organism. For a bacterium containing a single chromosome, a genome project will aim to map the sequence of that chromosome. For the human species, whose genome includes 22 pairs of autosomes and 2 sex chromosomes, a complete genome sequence will involve 46 separate chromosome sequences.

Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural landmarks. In this branch of genomics, whole or large parts of genomes resulting from genome projects are compared to study basic biological similarities and differences as well as evolutionary relationships between organisms. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA that is evolutionarily conserved between them. Therefore, comparative genomic approaches start with making some form of alignment of genome sequences and looking for orthologous sequences in the aligned genomes and checking to what extent those sequences are conserved. Based on these, genome and molecular evolution are inferred and this may in turn be put in the context of, for example, phenotypic evolution or population genetics.
C-value is the amount, in picograms, of DNA contained within a haploid nucleus or one half the amount in a diploid somatic cell of a eukaryotic organism. In some cases, the terms C-value and genome size are used interchangeably; however, in polyploids the C-value may represent two or more genomes contained within the same nucleus. Greilhuber et al. have suggested some new layers of terminology and associated abbreviations to clarify this issue, but these somewhat complex additions are yet to be used by other authors.

Genome size is the total amount of DNA contained within one copy of a single complete genome. It is typically measured in terms of mass in picograms or less frequently in daltons, or as the total number of nucleotide base pair ed Mb or Mbp). One picogram is equal to 978 megabases. In diploid organisms, genome size is often used interchangeably with the term C-value.
T. Ryan Gregory is a Canadian evolutionary biologist and genome biologist and a Professor and Chair of the Department of Integrative Biology and the Biodiversity Institute of Ontario at the University of Guelph in Guelph, Ontario, Canada.
The Plant DNA C-values Database is a comprehensive catalogue of C-value data for land plants and algae. The database was created by Prof. Michael D. Bennett and Dr. Ilia J. Leitch of the Royal Botanic Gardens, Kew, UK. The database was originally launched as the "Angiosperm DNA C-values Database" in April 1997, essentially as an online version of collected data lists that had been published by Prof. Bennett and colleagues since the 1970s. Release 1.0 of the more inclusive Plant DNA C-values Database was launched in 2001, with subsequent releases 2.0 in January 2003 and 3.0 in December 2004. In addition to the angiosperm dataset made available in 1997, the database has been expanded taxonomically several times and now includes data from pteridophytes, gymnosperms, bryophytes, and algae. .
A transgene is a gene that has been transferred naturally, or by any of a number of genetic engineering techniques from one organism to another. The introduction of a transgene, in a process known as transgenesis, has the potential to change the phenotype of an organism. Transgene describes a segment of DNA containing a gene sequence that has been isolated from one organism and is introduced into a different organism. This non-native segment of DNA may either retain the ability to produce RNA or protein in the transgenic organism or alter the normal function of the transgenic organism's genetic code. In general, the DNA is incorporated into the organism's germ line. For example, in higher vertebrates this can be accomplished by injecting the foreign DNA into the nucleus of a fertilized ovum. This technique is routinely used to introduce human disease genes or other genes of interest into strains of laboratory mice to study the function or pathology involved with that particular gene.
The Leibniz Institute DSMZ - German Collection of Microorganisms and Cell Cultures GmbH was founded 1969 as the national culture collection in Germany. This independent non-profit organization is dedicated to the acquisition, characterization, identification, preservation, distribution of Bacteria, Archea, fungi, plasmids, bacteriophages, human and animal cell lines, plant cell cultures and plant viruses. The organization is member of the German Wissenschaftsgemeinschaft Gottfried Wilhelm Leibniz and of worldwide organizations like the European Culture Collections' Organisation (ECCO), the World Federation for Culture Collections (WFCC), and the Global Biodiversity Information Facility (GBIF).

Plant genetics is the study of genes, genetic variation, and heredity specifically in plants. It is generally considered a field of biology and botany, but intersects frequently with many other life sciences and is strongly linked with the study of information systems. Plant genetics is similar in many ways to animal genetics but differs in a few key areas.

Genetic engineering is the science of manipulating genetic material of an organism. The first artificial genetic modification accomplished using biotechnology was transgenesis, the process of transferring genes from one organism to another, was first accomplished by Herbert Boyer and Stanley Cohen in 1973. It was the result of a series of advancements in techniques that allowed the direct modification of the genome. Important advances included the discovery of restriction enzymes and DNA ligases, the ability to design plasmids and technologies like polymerase chain reaction and sequencing. Transformation of the DNA into a host organism was accomplished with the invention of biolistics, Agrobacterium-mediated recombination and microinjection. The first genetically modified animal was a mouse created in 1974 by Rudolf Jaenisch. In 1976 the technology was commercialised, with the advent of genetically modified bacteria that produced somatostatin, followed by insulin in 1978. In 1983 an antibiotic resistant gene was inserted into tobacco, leading to the first genetically engineered plant. Advances followed that allowed scientists to manipulate and add genes to a variety of different organisms and induce a range of different effects. Plants were first commercialized with virus resistant tobacco released in China in 1992. The first genetically modified food was the Flavr Savr tomato marketed in 1994. By 2010, 29 countries had planted commercialized biotech crops. In 2000 a paper published in Science introduced golden rice, the first food developed with increased nutrient value.

Genetic engineering can be accomplished using multiple techniques. There are a number of steps that are followed before a genetically modified organism]] (GMO) is created. Genetic engineers must first choose what gene they wish to insert, modify, or delete. The gene must then be isolated and incorporated, along with other genetic elements, into a suitable Vector molecular biology vector. This vector is then used to insert the gene into the host genome, creating a transgenic or edited organism. The ability to genetically engineer organisms is built on years of research and discovery on how genes function and how we can manipulate them. Important advances included the discovery of restriction enzymes and DNA ligase is and the development of polymerase chain reaction]] and sequencing.
The onion test is a way of assessing the validity of an argument for a functional role for non-coding DNA, sometimes called "junk DNA". It relates to the paradox that would emerge if the majority of eukaryotic non-coding DNA would be assumed to be functional and the difficulty of reconciling that assumption with the diversity in genome sizes among species. The term "onion test" was originally proposed informally in a blog post by T. Ryan Gregory in order to help clarify the debate about junk DNA. The term has been mentioned in newspapers and online media, scientific journal articles, and a textbook. The test is defined as:
The onion test is a simple reality check for anyone who thinks they have come up with a universal function for junk DNA. Whatever your proposed function, ask yourself this question: Can I explain why an onion needs about five times more non-coding DNA for this function than a human?
The G-value paradox arises from the lack of correlation between the number of protein-coding genes among eukaryotes and their relative biological complexity. The microscopic nematode Caenorhabditis elegans, for example, is composed of only a thousand cells but has about the same number of genes as a human. Researchers suggest resolution of the paradox may lie in mechanisms such as alternative splicing and complex gene regulation that make the genes of humans and other complex eukaryotes relatively more productive.








