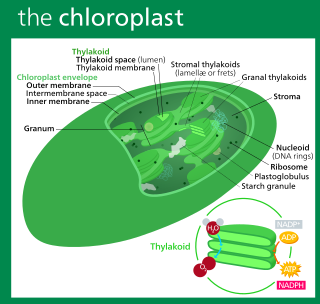
A chloroplast is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it in the energy-storage molecules ATP and NADPH while freeing oxygen from water in the cells. The ATP and NADPH is then used to make organic molecules from carbon dioxide in a process known as the Calvin cycle. Chloroplasts carry out a number of other functions, including fatty acid synthesis, amino acid synthesis, and the immune response in plants. The number of chloroplasts per cell varies from one, in unicellular algae, up to 100 in plants like Arabidopsis and wheat.
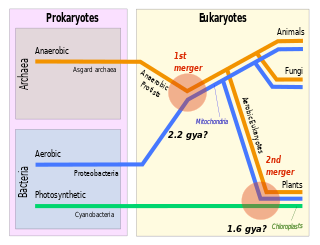
Symbiogenesis is the leading evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms. The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells are descended from formerly free-living prokaryotes taken one inside the other in endosymbiosis. Mitochondria appear to be phylogenetically related to Rickettsiales bacteria, while chloroplasts are thought to be related to cyanobacteria.
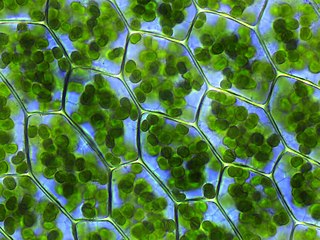
A plastid, pl. plastids, is a membrane-bound organelle found in the cells of plants, algae, and some other eukaryotic organisms. They are considered to be intracellular endosymbiotic cyanobacteria.

Chlamydomonas is a genus of green algae consisting of about 150 species of unicellular flagellates, found in stagnant water and on damp soil, in freshwater, seawater, and even in snow as "snow algae". Chlamydomonas is used as a model organism for molecular biology, especially studies of flagellar motility and chloroplast dynamics, biogenesis, and genetics. One of the many striking features of Chlamydomonas is that it contains ion channels (channelrhodopsins) that are directly activated by light. Some regulatory systems of Chlamydomonas are more complex than their homologs in Gymnosperms, with evolutionarily related regulatory proteins being larger and containing additional domains.

Pyrenoids are sub-cellular micro-compartments found in chloroplasts of many algae, and in a single group of land plants, the hornworts. Pyrenoids are associated with the operation of a carbon-concentrating mechanism (CCM). Their main function is to act as centres of carbon dioxide (CO2) fixation, by generating and maintaining a CO2 rich environment around the photosynthetic enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). Pyrenoids therefore seem to have a role analogous to that of carboxysomes in cyanobacteria.
A hydrogenase is an enzyme that catalyses the reversible oxidation of molecular hydrogen (H2), as shown below:

Biohydrogen is H2 that is produced biologically. Interest is high in this technology because H2 is a clean fuel and can be readily produced from certain kinds of biomass, including biological waste. Furthermore some photosynthetic microorganisms are capable to produce H2 directly from water splitting using light as energy source.

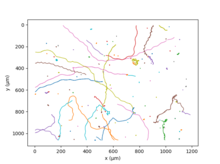
The eyespot apparatus is a photoreceptive organelle found in the flagellate or (motile) cells of green algae and other unicellular photosynthetic organisms such as euglenids. It allows the cells to sense light direction and intensity and respond to it, prompting the organism to either swim towards the light, or away from it. A related response occurs when cells are briefly exposed to high light intensity, causing the cell to stop, briefly swim backwards, then change swimming direction. Eyespot-mediated light perception helps the cells in finding an environment with optimal light conditions for photosynthesis. Eyespots are the simplest and most common "eyes" found in nature, composed of photoreceptors and areas of bright orange-red red pigment granules. Signals relayed from the eyespot photoreceptors result in alteration of the beating pattern of the flagella, generating a phototactic response.

In enzymology, ferredoxin hydrogenase, also referred to as [Fe-Fe]hydrogenase, H2 oxidizing hydrogenase, H2 producing hydrogenase, bidirectional hydrogenase, hydrogenase (ferredoxin), hydrogenlyase, and uptake hydrogenase, is found in Clostridium pasteurianum, Clostridium acetobutylicum,Chlamydomonas reinhardtii, and other organisms. The systematic name of this enzyme is hydrogen:ferredoxin oxidoreductase
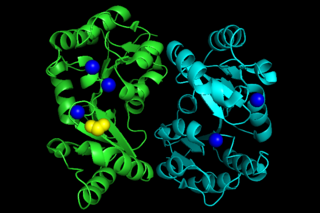
Phosphoglycolate phosphatase(EC 3.1.3.18; systematic name 2-phosphoglycolate phosphohydrolase), also commonly referred to as phosphoglycolate hydrolase, 2-phosphoglycolate phosphatase, P-glycolate phosphatase, and phosphoglycollate phosphatase, is an enzyme responsible for catalyzing the conversion of 2-phosphoglycolate into glycolate and phosphate:

Protochlorophyllide, or monovinyl protochlorophyllide, is an intermediate in the biosynthesis of chlorophyll a. It lacks the phytol side-chain of chlorophyll and the reduced pyrrole in ring D. Protochlorophyllide is highly fluorescent; mutants that accumulate it glow red if irradiated with blue light. In angiosperms, the later steps which convert protochlorophyllide to chlorophyll are light-dependent, and such plants are pale (chlorotic) if grown in the darkness. Gymnosperms, algae, and photosynthetic bacteria have another, light-independent enzyme and grow green in the darkness as well.

Phototaxis is a kind of taxis, or locomotory movement, that occurs when a whole organism moves towards or away from a stimulus of light. This is advantageous for phototrophic organisms as they can orient themselves most efficiently to receive light for photosynthesis. Phototaxis is called positive if the movement is in the direction of increasing light intensity and negative if the direction is opposite.

Guillardia is a genus of marine biflagellate cryptomonad algae with a plastid obtained through secondary endosymbiosis of a red alga.

Cyanidioschyzon merolae is a small (2μm), club-shaped, unicellular haploid red alga adapted to high sulfur acidic hot spring environments. The cellular architecture of C. merolae is extremely simple, containing only a single chloroplast and a single mitochondrion and lacking a vacuole and cell wall. In addition, the cellular and organelle divisions can be synchronized. For these reasons, C. merolae is considered an excellent model system for study of cellular and organelle division processes, as well as biochemistry and structural biology. The organism's genome was the first full algal genome to be sequenced in 2004; its plastid was sequenced in 2000 and 2003, and its mitochondrion in 1998. The organism has been considered the simplest of eukaryotic cells for its minimalist cellular organization.

Volvox carteri is a species of colonial green algae in the order Volvocales. The V. carteri life cycle includes a sexual phase and an asexual phase. V. carteri forms small spherical colonies, or coenobia, of 2000–6000 Chlamydomonas-type somatic cells and 12–16 large, potentially immortal reproductive cells called gonidia. While vegetative, male and female colonies are indistinguishable; however, in the sexual phase, females produce 35-45 eggs and males produce up to 50 sperm packets with 64 or 128 sperm each.
The D66 strain of Chlamydomonas reinhardtii, a single-celled green alga, is a cell-wall-deficient strain of algae that exhibits normal photosynthetic characteristics, but requires ammonia as a source of nitrogen for growth. This strain of green algae is becoming an increasingly popular research organism due to its potential to be used as a source of biofuels. The D66 strain's potential to produce clean and renewable biofuel has also made it an increasingly important topic in the field of conservation biology.

Chlororespiration is a respiratory process that takes place within plants. Inside plant cells there is an organelle called the chloroplast which is surrounded by the thylakoid membrane. This membrane contains an enzyme called NAD(P)H dehydrogenase which transfers electrons in a linear chain to oxygen molecules. This electron transport chain (ETC) within the chloroplast also interacts with those in the mitochondria where respiration takes place. Photosynthesis is also a process that Chlororespiration interacts with. If photosynthesis is inhibited by environmental stressors like water deficit, increased heat, and/or increased/decreased light exposure, or even chilling stress then chlororespiration is one of the crucial ways that plants use to compensate for chemical energy synthesis.
Sabeeha Sabanali Merchant is a professor of plant biology at the University of California, Berkeley. She studies the photosynthetic metabolism and metalloenzymes In 2010 Merchant led the team that sequenced the Chlamydomonas genome. She was elected a member of the National Academy of Sciences in 2012.

Protists are the eukaryotes that cannot be classified as plants, fungi or animals. They are mostly unicellular and microscopic. Many unicellular protists, particularly protozoans, are motile and can generate movement using flagella, cilia or pseudopods. Cells which use flagella for movement are usually referred to as flagellates, cells which use cilia are usually referred to as ciliates, and cells which use pseudopods are usually referred to as amoeba or amoeboids. Other protists are not motile, and consequently have no built-in movement mechanism.
Christoph Benning is a German–American plant biologist. He is an MSU Foundation Professor and University Distinguished Professor at Michigan State University. Benning's research into lipid metabolism in plants, algae and photosynthetic bacteria, led him to be named Editor-in-Chief of The Plant Journal in October 2008.