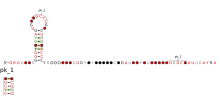An inverted repeat is a single stranded sequence of nucleotides followed downstream by its reverse complement. The intervening sequence of nucleotides between the initial sequence and the reverse complement can be any length including zero. For example, 5'---TTACGnnnnnnCGTAA---3' is an inverted repeat sequence. When the intervening length is zero, the composite sequence is a palindromic sequence.

In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3′-UTR often contains regulatory regions that post-transcriptionally influence gene expression.
The 5′ untranslated region is the region of an mRNA that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming complex secondary structure to regulate translation.
This is a list of topics in molecular biology. See also index of biochemistry articles.

Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions.

Antizyme RNA frameshifting stimulation element is a structural element which is found in antizyme mRNA and is known to promote frameshifting. Antizyme genes have two partially overlapping open reading frames, the second, which encodes the functional (antizyme) protein requires +1 translational frameshifting. This frameshift is stimulated by a pseudoknot present 3' of the frameshift site in the antizyme mRNA. The frameshifting efficiency is dependent on the concentration of polyamines in the cell, when the polyamine concentration is high frameshifting is more likely to occur which leads to an increase in the quantity of functional antizyme produced. The functional antizyme acts to reduce ornithine decarboxylase (ODC) activity which leads to a drop in polyamines present in the cell. Therefore, this family can be thought of as a biosensor for intracellular free polyamines that functions via a negative feedback loop.
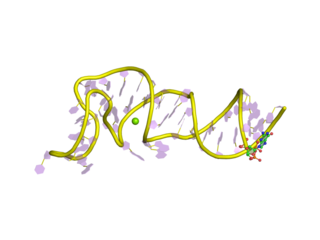
The Coronavirus 3′ stem-loop II-like motif is a secondary structure motif identified in the 3′ untranslated region (3′UTR) of astrovirus, coronavirus and equine rhinovirus genomes. Its function is unknown, but various viral 3′ UTR regions have been found to play roles in viral replication and packaging.

The Coronavirus 3′ UTR pseudoknot is an RNA structure found in the coronavirus genome. Coronaviruses contain 30 kb single-stranded positive-sense RNA genomes. The 3′ UTR region of these coronavirus genomes contains a conserved ~55 nucleotide pseudoknot structure which is necessary for viral genome replication. The mechanism of cis-regulation is unclear, but this element is postulated to function in the plus-strand.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Betacoronavirus. It has an important role in regulating the packaging of the viral genome into the capsid. As part of the viral life cycle, within the infected cell, the viral genome becomes associated with viral proteins and assembles into new infective progeny viruses. This process is called packaging and is vital for viral replication.

The PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine (Q) from GTP. PreQ1 (pre-queuosine1) is an intermediate in the queuosine pathway, and preQ1 riboswitch, as a type of riboswitch, is an RNA element that binds preQ1. The preQ1 riboswitch is distinguished by its unusually small aptamer, compared to other riboswitches. Its atomic-resolution three-dimensional structure has been determined, with the PDB ID 2L1V.

HIV ribosomal frameshift signal is a ribosomal frameshift (PRF) that human immunodeficiency virus (HIV) uses to translate several different proteins from the same sequence.
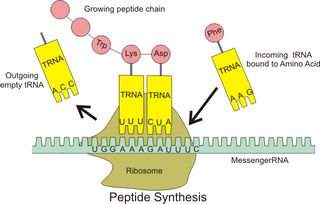
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses, retrotransposons and bacterial insertion elements, and also in some cellular genes.
The MAEB RNA motif is a conserved stem-loop RNA structure present in many species in the genus Burkholderia. MAEB stem-loops typically occur in blocks of repeats, usually with 2–6 consecutive instances of MAEB stem-loops separated by a short and conserved linker sequence. As many as 12 consecutive MAEB stem-loops have been observed in a single block.
ALIL pseudoknot is an RNA element that induces frameshifting in bacteria. The expression of a minority of genes requires frameshifting to occur where the frequency of frameshifting is increased by a RNA secondary structure located on the 3' side of the shift site. This structure can be either a pseudoknot or a stem-loop and acts as a physical barrier to mRNA translocation so therefore causes ribosome pausing.
Ribosomal pause refers to the queueing or stacking of ribosomes during translation of the nucleotide sequence of mRNA transcripts. These transcripts are decoded and converted into an amino acid sequence during protein synthesis by ribosomes. Due to the pause sites of some mRNA’s, there is a disturbance caused in translation. Ribosomal pausing occurs in both eukaryotes and prokaryotes.

A slippery sequence is a small section of codon nucleotide sequences that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosome to "slip." This allows a tRNA to shift by 1 base (−1) after it has paired with its anticodon, changing the reading frame. A −1 frameshift triggered by such a sequence is a Programmed −1 Ribosomal Frameshift. It is followed by a spacer region, and an RNA secondary structure. Such sequences are common in virus polyproteins.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 5′ end which is called the 5′ UTR. The 5′ UTR is responsible for important biological functions, such as viral replication, transcription and packaging. The 5′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 3′ end which is called the 3′ UTR. The 3′ UTR is responsible for important biological functions, such as viral replication. The 3′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.
Flavivirus 3' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.