Related Research Articles
An anaerobic organism or anaerobe is any organism that does not require molecular oxygen for growth. It may react negatively or even die if free oxygen is present. In contrast, an aerobic organism (aerobe) is an organism that requires an oxygenated environment. Anaerobes may be unicellular or multicellular. Most fungi are obligate aerobes, requiring oxygen to survive. However, some species, such as the Chytridiomycota that reside in the rumen of cattle, are obligate anaerobes; for these species, anaerobic respiration is used because oxygen will disrupt their metabolism or kill them. Deep waters of the ocean are a common anoxic environment.

The human microbiome is the aggregate of all microbiota that reside on or within human tissues and biofluids along with the corresponding anatomical sites in which they reside, including the skin, mammary glands, seminal fluid, uterus, ovarian follicles, lung, saliva, oral mucosa, conjunctiva, biliary tract, and gastrointestinal tract. Types of human microbiota include bacteria, archaea, fungi, protists, and viruses. Though micro-animals can also live on the human body, they are typically excluded from this definition. In the context of genomics, the term human microbiome is sometimes used to refer to the collective genomes of resident microorganisms; however, the term human metagenome has the same meaning.

The Bacillota are a phylum of bacteria, most of which have gram-positive cell wall structure. The renaming of phyla such as Firmicutes in 2021 remains controversial among microbiologists, many of whom continue to use the earlier names of long standing in the literature.

Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics.

Gut microbiota, gut microbiome, or gut flora, are the microorganisms, including bacteria, archaea, fungi, and viruses, that live in the digestive tracts of animals. The gastrointestinal metagenome is the aggregate of all the genomes of the gut microbiota. The gut is the main location of the human microbiome. The gut microbiota has broad impacts, including effects on colonization, resistance to pathogens, maintaining the intestinal epithelium, metabolizing dietary and pharmaceutical compounds, controlling immune function, and even behavior through the gut–brain axis.
Dysbiosis is characterized by a disruption to the microbiome resulting in an imbalance in the microbiota, changes in their functional composition and metabolic activities, or a shift in their local distribution. For example, a part of the human microbiota such as the skin flora, gut flora, or vaginal flora, can become deranged, with normally dominating species underrepresented and normally outcompeted or contained species increasing to fill the void. Dysbiosis is most commonly reported as a condition in the gastrointestinal tract.

The Human Microbiome Project (HMP) was a United States National Institutes of Health (NIH) research initiative to improve understanding of the microbiota involved in human health and disease. Launched in 2007, the first phase (HMP1) focused on identifying and characterizing human microbiota. The second phase, known as the Integrative Human Microbiome Project (iHMP) launched in 2014 with the aim of generating resources to characterize the microbiome and elucidating the roles of microbes in health and disease states. The program received $170 million in funding by the NIH Common Fund from 2007 to 2016.

Microbiota are the range of microorganisms that may be commensal, mutualistic, or pathogenic found in and on all multicellular organisms, including plants. Microbiota include bacteria, archaea, protists, fungi, and viruses, and have been found to be crucial for immunologic, hormonal, and metabolic homeostasis of their host.
Metaproteomics is an umbrella term for experimental approaches to study all proteins in microbial communities and microbiomes from environmental sources. Metaproteomics is used to classify experiments that deal with all proteins identified and quantified from complex microbial communities. Metaproteomics approaches are comparable to gene-centric environmental genomics, or metagenomics.
Alistipes is a Gram-negative genus of rod-shaped anaerobic bacteria in the phylum Bacteroidota. When members of this genus colonize the human gastrointestinal (GI) tract, they provide protective effects against colitis, autism, and cirrhosis. However, this genus can also cause dysbiosis by contributing to anxiety, chronic fatigue syndrome, depression, and hypertension. Showcasing priority effects in microbiome assembly, when infant GI tracts have bacteria of the species Staphylococcus but not the species Faecalibacterium, Alistipes species become less capable of colonization.
Microbial phylogenetics is the study of the manner in which various groups of microorganisms are genetically related. This helps to trace their evolution. To study these relationships biologists rely on comparative genomics, as physiology and comparative anatomy are not possible methods.
Sutterella is a genus of Gram-negative, rod-shaped, non-spore-forming, Betaproteobacteria whose species have been isolated from the human gastrointestinal tract as well as canine feces. The genus of the family Sutterellaceae currently encompasses 4 distinct species, though at least 5 additional species have been proposed that do not yet meet International Code of Nomenclature of Prokaryotes (ICNP) standards for classification. Sutterella are frequently referred to as commensal in the context of human hosts, but are associated with inflammation, which has implications for a number of diseases.

Akkermansia muciniphila is a human intestinal symbiont, isolated from human feces. It is a mucin-degrading bacterium belonging to the genus, Akkermansia, discovered in 2004 by Muriel Derrien and Willem de Vos at Wageningen University of the Netherlands. It belongs to the phylum Verrucomicrobiota and its type strain is MucT. It is under preliminary research for its potential association with metabolic disorders.
Methanobrevibacter oralis is a methanogenic archaeon species considered to be a member of the human microbiota, mainly associated to the oral cavity. M. oralis is a coccobacillary shaped, single-cell, Gram-positive, non-motile microorganism of the Archaea domain of life. This species has been isolated and sequenced from humans in dental plaque and in their gastrointestinal tract. As a methanogen and a hydrogenotroph, this prokaryote can produce methane by using hydrogen and carbon dioxide as substrates through a process called methanogenesis.
Mark J. Pallen is a research leader at the Quadram Institute and Professor of Microbial Genomics at the University of East Anglia. In recent years, he has been at the forefront of efforts to apply next-generation sequencing to problems in microbiology and ancient DNA research.

A microbiome is the community of microorganisms that can usually be found living together in any given habitat. It was defined more precisely in 1988 by Whipps et al. as "a characteristic microbial community occupying a reasonably well-defined habitat which has distinct physio-chemical properties. The term thus not only refers to the microorganisms involved but also encompasses their theatre of activity". In 2020, an international panel of experts published the outcome of their discussions on the definition of the microbiome. They proposed a definition of the microbiome based on a revival of the "compact, clear, and comprehensive description of the term" as originally provided by Whipps et al., but supplemented with two explanatory paragraphs. The first explanatory paragraph pronounces the dynamic character of the microbiome, and the second explanatory paragraph clearly separates the term microbiota from the term microbiome.
Metatranscriptomics is the set of techniques used to study gene expression of microbes within natural environments, i.e., the metatranscriptome.
Hologenomics is the omics study of hologenomes. A hologenome is the whole set of genomes of a holobiont, an organism together with all co-habitating microbes, other life forms, and viruses. While the term hologenome originated from the hologenome theory of evolution, which postulates that natural selection occurs on the holobiont level, hologenomics uses an integrative framework to investigate interactions between the host and its associated species. Examples include gut microbe or viral genomes linked to human or animal genomes for host-microbe interaction research. Hologenomics approaches have also been used to explain genetic diversity in the microbial communities of marine sponges.
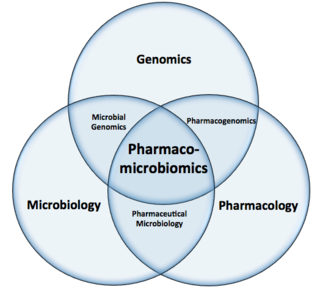
Pharmacomicrobiomics, proposed by Prof. Marco Candela for the ERC-2009-StG project call, and publicly coined for the first time in 2010 by Rizkallah et al., is defined as the effect of microbiome variations on drug disposition, action, and toxicity. Pharmacomicrobiomics is concerned with the interaction between xenobiotics, or foreign compounds, and the gut microbiome. It is estimated that over 100 trillion prokaryotes representing more than 1000 species reside in the gut. Within the gut, microbes help modulate developmental, immunological and nutrition host functions. The aggregate genome of microbes extends the metabolic capabilities of humans, allowing them to capture nutrients from diverse sources. Namely, through the secretion of enzymes that assist in the metabolism of chemicals foreign to the body, modification of liver and intestinal enzymes, and modulation of the expression of human metabolic genes, microbes can significantly impact the ingestion of xenobiotics.
Sediminibacillus is a genus of bacteria from the family of Bacillaceae. Sediminibacillus species are halophilic bacteria and found in salty human stools and marine sponges. Sediminibacillus species are identified from Plakortis dariae sponge of the Saint Martin's island of the Bay of Bengal, Bangladesh.
References
- ↑ Lagier J, Armougom F, Million M, et al. (December 2012). "Microbial culturomics: paradigm shift in the human gut microbiome study". Clinical Microbiology and Infection. 18 (12): 1185–1193. doi: 10.1111/1469-0691.12023 .
- 1 2 Lagier J, Khelaifia S, Alou M, et al. (December 2016). "Culture of previously uncultured members of the human gut microbiota by culturomics". Nature Microbiology. 1 (12): 16203. doi: 10.1038/nmicrobiol.2016.203 .
- 1 2 3 Greub, G. (December 2012). "Culturomics: a new approach to study the human microbiome". Clinical Microbiology and Infection. 18 (12): 1157–1159. doi: 10.1111/1469-0691.12032 .
- ↑ Rinke C, Schwientek P, Sczyrba A, et al. (25 July 2013). "Insights into the phylogeny and coding potential of microbial dark matter". Nature. 499 (7459): 431–437. doi: 10.1038/nature12352 . hdl: 10453/27467 .
- ↑ Diakite A, Dubourg G, Dione N, et al. (December 2020). "Optimization and standardization of the culturomics technique for human microbiome exploration". Scientific Reports. 10 (1): 9674. doi: 10.1038/s41598-020-66738-8 . PMC 7295790 .
- ↑ Chang Y, Hou F, Pan Z, et al. (17 December 2019). "Optimization of culturomics strategy in human fecal samples". Frontiers in Microbiology. 10: 2891. doi: 10.3389/fmicb.2019.02891 . PMC 6927924 .
- ↑ Diakite A, Dubourg G, Dione N, et al. (21 October 2019). "Extensive culturomics of 8 healthy samples enhances metagenomics efficiency". PLOS ONE. 14 (10): e0223543. doi: 10.1371/journal.pone.0223543 . PMC 6802823 .