
The Epstein–Barr virus (EBV), formally called Human gammaherpesvirus 4, is one of the nine known human herpesvirus types in the herpes family, and is one of the most common viruses in humans. EBV is a double-stranded DNA virus. Epstein–Barr virus (EBV) is the first identified oncogenic virus, which establishes permanent infection in humans. EBV causes infectious mononucleosis and is also tightly linked to many malignant diseases. Various vaccine formulations underwent testing in different animals or in humans. However, none of them were able to prevent EBV infection and no vaccine has been approved to date.
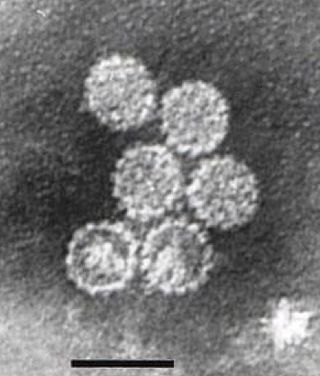
Papillomaviridae is a family of non-enveloped DNA viruses whose members are known as papillomaviruses. Several hundred species of papillomaviruses, traditionally referred to as "types", have been identified infecting all carefully inspected mammals, but also other vertebrates such as birds, snakes, turtles and fish. Infection by most papillomavirus types, depending on the type, is either asymptomatic or causes small benign tumors, known as papillomas or warts. Papillomas caused by some types, however, such as human papillomaviruses 16 and 18, carry a risk of becoming cancerous.
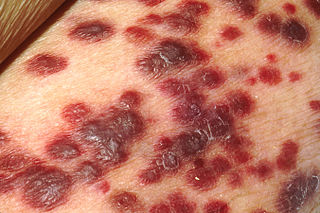
Kaposi's sarcoma-associated herpesvirus (KSHV) is the ninth known human herpesvirus; its formal name according to the International Committee on Taxonomy of Viruses (ICTV) is Human gammaherpesvirus 8, or HHV-8 in short. Like other herpesviruses, its informal names are used interchangeably with its formal ICTV name. This virus causes Kaposi's sarcoma, a cancer commonly occurring in AIDS patients, as well as primary effusion lymphoma, HHV-8-associated multicentric Castleman's disease and KSHV inflammatory cytokine syndrome. It is one of seven currently known human cancer viruses, or oncoviruses. Even after many years since the discovery of KSHV/HHV8, there is no known cure for KSHV associated tumorigenesis.

Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome and packaging these copies, the virus continues infecting new hosts. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm.

Adeno-associated viruses (AAV) are small viruses that infect humans and some other primate species. They belong to the genus Dependoparvovirus, which in turn belongs to the family Parvoviridae. They are small replication-defective, nonenveloped viruses and have linear single-stranded DNA (ssDNA) genome of approximately 4.8 kilobases (kb).
Virus latency is the ability of a pathogenic virus to lie dormant within a cell, denoted as the lysogenic part of the viral life cycle. A latent viral infection is a type of persistent viral infection which is distinguished from a chronic viral infection. Latency is the phase in certain viruses' life cycles in which, after initial infection, proliferation of virus particles ceases. However, the viral genome is not eradicated. The virus can reactivate and begin producing large amounts of viral progeny without the host becoming reinfected by new outside virus, and stays within the host indefinitely.

Human herpesvirus 6 (HHV-6) is the common collective name for human betaherpesvirus 6A (HHV-6A) and human betaherpesvirus 6B (HHV-6B). These closely related viruses are two of the nine known herpesviruses that have humans as their primary host.

Herpesviridae is a large family of DNA viruses that cause infections and certain diseases in animals, including humans. The members of this family are also known as herpesviruses. The family name is derived from the Greek word ἕρπειν, referring to spreading cutaneous lesions, usually involving blisters, seen in flares of herpes simplex 1, herpes simplex 2 and herpes zoster (shingles). In 1971, the International Committee on the Taxonomy of Viruses (ICTV) established Herpesvirus as a genus with 23 viruses among four groups. As of 2020, 115 species are recognized, all but one of which are in one of the three subfamilies. Herpesviruses can cause both latent and lytic infections.

Human Herpes Virus (HHV) Infected Cell Polypeptide 0 (ICP0) is a protein, encoded by the DNA of herpes viruses. It is produced by herpes viruses during the earliest stage of infection, when the virus has recently entered the host cell; this stage is known as the immediate-early or α ("alpha") phase of viral gene expression. During these early stages of infection, ICP0 protein is synthesized and transported to the nucleus of the infected host cell. Here, ICP0 promotes transcription from viral genes, disrupts structures in the nucleus known as nuclear dots or promyelocytic leukemia (PML) nuclear bodies, and alters the expression of host and viral genes in combination with a neuron specific protein. At later stages of cellular infection, ICP0 relocates to the cell cytoplasm to be incorporated into new virion particles.

Herpes simplex virus1 and 2, also known by their taxonomic names Human alphaherpesvirus 1 and Human alphaherpesvirus 2, are two members of the human Herpesviridae family, a set of viruses that produce viral infections in the majority of humans. Both HSV-1 and HSV-2 are very common and contagious. They can be spread when an infected person begins shedding the virus.

Gammaherpesvirinae is a subfamily of viruses in the order Herpesvirales and in the family Herpesviridae. Viruses in Gammaherpesvirinae are distinguished by reproducing at a more variable rate than other subfamilies of Herpesviridae. Mammals serve as natural hosts. There are 43 species in this subfamily, divided among 7 genera with three species unassigned to a genus. Diseases associated with this subfamily include: HHV-4: infectious mononucleosis. HHV-8: Kaposi's sarcoma.
HHV Capsid Portal Protein, or HSV-1 UL-6 protein, is the protein which forms a cylindrical portal in the capsid of Herpes simplex virus (HSV-1). The protein is commonly referred to as the HSV-1 UL-6 protein because it is the transcription product of Herpes gene UL-6.
The Epstein–Barr virus nuclear antigen 2 (EBNA-2) is one of the six EBV viral nuclear proteins expressed in latently infected B lymphocytes is a transactivator protein. EBNA2 is involved in the regulation of latent viral transcription and contributes to the immortalization of EBV infected cells. EBNA2 acts as an adapter molecule that binds to cellular sequence-specific DNA-binding proteins, JK recombination signal-binding protein (RBP-JK), and PU.1 as well as working with multiple members of the RNA polymerase II transcription complex.
Human betaherpesvirus 7 (HHV-7) is one of nine known members of the Herpesviridae family that infects humans. HHV-7 is a member of Betaherpesvirinae, a subfamily of the Herpesviridae that also includes HHV-6 and Cytomegalovirus. HHV-7 often acts together with HHV-6, and the viruses together are sometimes referred to by their genus, Roseolovirus. HHV-7 was first isolated in 1990 from CD4+ T cells taken from peripheral blood lymphocytes.
ICP8, the herpes simplex virus type-1 single-strand DNA-binding protein, is one of seven proteins encoded in the viral genome of HSV-1 that is required for HSV-1 DNA replication. It is able to anneal to single-stranded DNA (ssDNA) as well as melt small fragments of double-stranded DNA (dsDNA); its role is to destabilize duplex DNA during initiation of replication. It differs from helicases because it is ATP- and Mg2+-independent. In cells infected with HSV-1, the DNA in those cells become colocalized with ICP8.
David Mahan Knipe is the Higgins Professor of Microbiology and Molecular Genetics in the Department of Microbiology at the Harvard Medical School in Boston, Massachusetts and co-chief editor of the reference book Fields Virology. He returned to the Chair of the Program in Virology at Harvard Medical School in 2019, having previously held the position from 2004 through 2016 and served as interim Co-Chair of the Microbiology and Immunobiology Department from 2016 through 2018.
BZLF1, also known as Zta, EB1, is an immediate-early viral gene of the Epstein–Barr virus (EBV) of the Herpes Virus Family, which induces cancers and infects primarily the B-cells of 95% of the human population. This gene produces the expression of other EBV genes in other stages of disease progression, and is involved in converting the virus from the latent to the lytic form.
Human herpes viruses, also known as HHVs, are part of a family of DNA viruses that cause several diseases in humans. One of the most notable functions of this virus family is their ability to enter a latent phase and lay dormant within animals for extended periods of time. The mechanism that controls this is very complex because expression of viral proteins during latency is decreased a great deal, meaning that the virus must have transcription of its genes repressed. There are many factors and mechanisms that control this process and epigenetics is one way this is accomplished. Epigenetics refers to persistent changes in expression patterns that are not caused by changes to the DNA sequence. This happens through mechanisms such as methylation and acetylation of histones, DNA methylation, and non-coding RNAs (ncRNA). Altering the acetylation of histones creates changes in expression by changing the binding affinity of histones to DNA, making it harder or easier for transcription machinery to access the DNA. Methyl and acetyl groups can also act as binding sites for transcription factors and enzymes that further modify histones or alter the DNA itself.
HSV epigenetics is the epigenetic modification of herpes simplex virus (HSV) genetic code.
Human Immunodeficiency Virus (HIV) has the capability to enter a latent stage of infection where it exists as a dormant provirus in CD4+ T-cells. Most latently infected cells are resting memory T cells, however a small fraction of latently infected cells isolated from HIV patients are naive CD4 T cells.