Related Research Articles
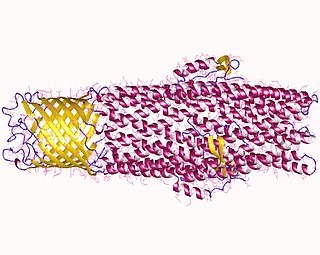
In microbiology, efflux is the moving of a variety of different compounds out of cells, such as antibiotics, heavy metals, organic pollutants, plant-produced compounds, quorum sensing signals, bacterial metabolites and neurotransmitters. All microorganisms, with a few exceptions, have highly conserved DNA sequences in their genome that encode efflux pumps. Efflux pumps actively move substances out of a microorganism, in a process known as active efflux, which is a vital part of xenobiotic metabolism. This active efflux mechanism is responsible for various types of resistance to bacterial pathogens within bacterial species - the most concerning being antibiotic resistance because microorganisms can have adapted efflux pumps to divert toxins out of the cytoplasm and into extracellular media.
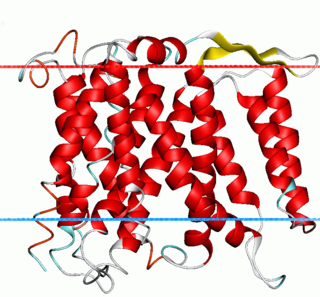
The sodium–hydrogen antiporter or sodium–proton exchanger (Na+/H+ exchanger) is a membrane protein that transports Na+ into the cell, and H+ out of the cell (antiport).
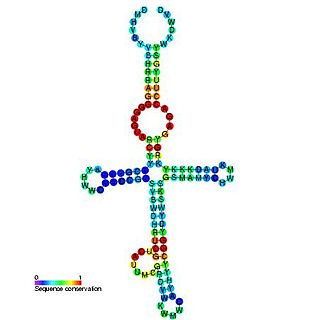
The yybP-ykoY leader RNA element was originally discovered in E. coli during a large scale screen and was named SraF. This family was later found to exist upstream of related families of protein genes in many bacteria, including the yybP and ykoY genes in B. subtilis. The specific functions of these proteins are unknown, but this structured RNA element may be involved in their genetic regulation as a riboswitch. The yybP-ykoY element was later proposed to be manganese-responsive after another associated family of genes, YebN/MntP, was shown to encode Mn2+ efflux pumps in several bacteria. Genetic data and a crystal structure confirmed that yybp-ykoY is a manganese riboswitch that directly binds Mn2+

The enzyme Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

Cystine/glutamate transporter is an antiporter that in humans is encoded by the SLC7A11 gene.

Multidrug and toxin extrusion protein 1 (MATE1), also known as solute carrier family 47 member 1, is a protein that in humans is encoded by the SLC47A1 gene. SLC47A1 belongs to the MATE family of transporters that are found in bacteria, archaea and eukaryotes.

Multidrug and toxin extrusion protein 2 is a protein which in humans is encoded by the SLC47A2 gene.
Multi-antimicrobial extrusion protein (MATE) also known as multidrug and toxin extrusion or multidrug and toxic compound extrusion is a family of proteins which function as drug/sodium or proton antiporters.
Small multidrug resistance protein is a family of integral membrane proteins that confer drug resistance to a wide range of toxic compounds by removing them for the cells. The efflux is coupled to an influx of protons. An example is Escherichia coli mvrC P23895 which prevents the incorporation of methyl viologen into cells and is involved in ethidium bromide efflux.
Cation diffusion facilitators (CDFs) are transmembrane proteins that provide tolerance of cells to divalent metal ions, such as cadmium, zinc, and cobalt. These proteins are considered to be efflux pumps that remove these divalent metal ions from cells. However, some members of the CDF superfamily are implicated in ion uptake. All members of the CDF family possess six putative transmembrane spanners with strongest conservation in the four N-terminal spanners. The Cation Diffusion Facilitator (CDF) Superfamily includes the following families:
The Amino Acid-Polyamine-Organocation (APC) Family of transport proteins includes members that function as solute:cation symporters and solute:solute antiporters. They occur in bacteria, archaea, fungi, unicellular eukaryotic protists, slime molds, plants and animals. They vary in length, being as small as 350 residues and as large as 850 residues. The smaller proteins are generally of prokaryotic origin while the larger ones are of eukaryotic origin. Most of them possess twelve transmembrane α-helical spanners but have a re-entrant loop involving TMSs 2 and 3. The APC Superfamily was established to encompass a wider range of homologues.
The potassium (K+) uptake permease (KUP) family (TC# 2.A.72) is a member of the APC superfamily of secondary carriers. Proteins of the KUP/HAK/KT family include the KUP (TrkD) protein of E. coli and homologues in both Gram-positive and Gram-negative bacteria. High affinity (20 μM) K+ uptake systems (Hak1, TC# 2.A.72.2.1) of the yeast Debaryomyces occidentalis as well as the fungus, Neurospora crassa, and several homologues in plants have been characterized. Arabidopsis thaliana and other plants possess multiple KUP family paralogues. While many plant proteins cluster tightly together, the Hak1 proteins from yeast as well as the two Gram-positive and Gram-negative bacterial proteins are distantly related on the phylogenetic tree for the KUP family. All currently classified members of the KUP family can be found in the Transporter Classification Database.
Lysine Exporters are a superfamily of transmembrane proteins which export amino acids, lipids and heavy metal ions. They provide ionic homeostasis, play a role in cell envelope assembly, and protect from excessive concentrations of heavy metals in cytoplasm. The superfamily was named based on the early discovery of the LysE carrier protein of Corynebacterium glutamicum.
The arsenical resistance-3 (ACR3) family is a member of the BART superfamily. Based on operon analyses, ARC3 homologues may function either as secondary carriers or as primary active transporters, similarly to the ArsB and ArsAB families. In the latter case ATP hydrolysis again energizes transport. ARC3 homologues transport the same anions as ArsA/AB homologues, though ArsB homologues are members of the IT Superfamily and homologues of the ARC3 family are within the BART Superfamily suggesting they may not be evolutionarily related.

Na+/H+ antiporter A (NhaA) family (TC# 2.A.33) contains a number of bacterial sodium-proton antiporter (SPAP) proteins. These are integral membrane proteins that catalyse the exchange of H+ for Na+ in a manner that is highly pH dependent. Homologues have been sequenced from a number of bacteria and archaea. Prokaryotes possess multiple paralogues. A representative list of the proteins that belong to the NhaA family can be found in the Transporter Classification Database.
The NhaB family belongs to the ion transporter (IT) superfamily. A representative list of proteins belonging to the NhaB family can be found in the Transporter Classification Database.
The NhaC family belongs to the Ion Transporter (IT) Superfamily. A representative list of proteins belonging to the NhaC family can be found in the Transporter Classification Database.

The Monovalent Cation:Proton Antiporter-1 (CPA1) Family (TC# 2.A.36) is a large family of proteins derived from Gram-positive and Gram-negative bacteria, blue-green bacteria, archaea, yeast, plants and animals. The CPA1 family belongs to the VIC superfamily. Transporters from eukaryotes have been functionally characterized to catalyze Na+:H+ exchange. Their primary physiological functions are thought to be in (1) cytoplasmic pH regulation, extruding the H+ generated during metabolism, and (2) salt tolerance (in plants), due to Na+ uptake into vacuoles. Bacterial homologues have also been found to facilitate Na+:H+ antiport, but some also catalyze Li+:H+ antiport or Ca2+:H+ antiport under certain conditions.
The Monovalent Cation (K+ or Na+):Proton Antiporter-3 (CPA3) Family (TC# 2.A.63) is a member of the Na+ transporting Mrp superfamily. The CPA3 family consists of bacterial multicomponent K+:H+ and Na+:H+ antiporters. The best characterized systems are the PhaABCDEFG system of Sinorhizobium meliloti (TC# 2.A.63.1.1) that functions in pH adaptation and as a K+ efflux system, and the MnhABCDEFG system of Staphylococcus aureus (TC# 2.A.63.1.3) that functions as a Na+ efflux Na+:H+ antiporter.
The cation:proton antiporter (CPA) superfamily is a superfamily of transport proteins named after one of its constituent members, the monovalent cation:proton antiporter-2 (CPA2).
References
- 1 2 Healy J, Ekkerman S, Pliotas C, Richard M, Bartlett W, Grayer SC, Morris GM, Miller S, Booth IR, Conway SJ, Rasmussen T (April 2014). "Understanding the structural requirements for activators of the Kef bacterial potassium efflux system". Biochemistry. 53 (12): 1982–92. doi:10.1021/bi5001118. PMC 4004266 . PMID 24601535.
- ↑ "2.A.37 The Monovalent Cation:Proton Antiporter-2 (CPA2) Family". Transporter Classification Database. Retrieved 2016-03-16.
- ↑ Bakker EP, Borchard A, Michels M, Altendorf K, Siebers A (September 1987). "High-affinity potassium uptake system in Bacillus acidocaldarius showing immunological cross-reactivity with the Kdp system from Escherichia coli". Journal of Bacteriology. 169 (9): 4342–8. doi:10.1128/jb.169.9.4342-4348.1987. PMC 213750 . PMID 2957359.
- 1 2 Booth, I.R.; Jones, M.A.; McLaggan, D; Nikolaev, Y (1996). Konings, W.N. (ed.). Bacterial Ion Channels. ISBN 978-0-444-82442-4.
{{cite book}}:|work=ignored (help) - ↑ Munro AW, Ritchie GY, Lamb AJ, Douglas RM, Booth IR (March 1991). "The cloning and DNA sequence of the gene for the glutathione-regulated potassium-efflux system KefC of Escherichia coli". Molecular Microbiology. 5 (3): 607–16. doi:10.1111/j.1365-2958.1991.tb00731.x. PMID 2046548. S2CID 23871816.
- ↑ Waser M, Hess-Bienz D, Davies K, Solioz M (March 1992). "Cloning and disruption of a putative NaH-antiporter gene of Enterococcus hirae". The Journal of Biological Chemistry. 267 (8): 5396–400. doi: 10.1016/S0021-9258(18)42779-2 . PMID 1312090.
- 1 2 Ferguson GP, Munro AW, Douglas RM, McLaggan D, Booth IR (September 1993). "Activation of potassium channels during metabolite detoxification in Escherichia coli". Molecular Microbiology. 9 (6): 1297–303. doi:10.1111/j.1365-2958.1993.tb01259.x. PMID 7934942. S2CID 26045169.
- 1 2 Ferguson GP, Nikolaev Y, McLaggan D, Maclean M, Booth IR (February 1997). "Survival during exposure to the electrophilic reagent N-ethylmaleimide in Escherichia coli: role of KefB and KefC potassium channels". Journal of Bacteriology. 179 (4): 1007–12. doi:10.1128/jb.179.4.1007-1012.1997. PMC 178791 . PMID 9023177.
- ↑ Miller S, Ness LS, Wood CM, Fox BC, Booth IR (November 2000). "Identification of an ancillary protein, YabF, required for activity of the KefC glutathione-gated potassium efflux system in Escherichia coli". Journal of Bacteriology. 182 (22): 6536–40. doi:10.1128/jb.182.22.6536-6540.2000. PMC 94807 . PMID 11053405.
- ↑ MacLean MJ, Ness LS, Ferguson GP, Booth IR (February 1998). "The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli". Molecular Microbiology. 27 (3): 563–71. doi: 10.1046/j.1365-2958.1998.00701.x . PMID 9489668. S2CID 10720918.
- ↑ Stumpe, S.; Schlösser, A.; Schleyer, M.; Bakker, E. P. (1996-01-01). "Chapter 21 K+ circulation across the prokaryotic cell membrane: K+-uptake systems". In W.N. Konings, H. R. Kaback and J. S. Lolkema (ed.). Transport Processes in Eukaryotic and Prokaryotic Organisms. Vol. 2. North-Holland. pp. 473–499. doi:10.1016/s1383-8121(96)80062-5. ISBN 9780444824424.
- ↑ Fujisawa M, Ito M, Krulwich TA (August 2007). "Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity". Proceedings of the National Academy of Sciences of the United States of America. 104 (33): 13289–94. Bibcode:2007PNAS..10413289F. doi: 10.1073/pnas.0703709104 . PMC 1948933 . PMID 17679694.
- 1 2 Roosild TP, Castronovo S, Healy J, Miller S, Pliotas C, Rasmussen T, Bartlett W, Conway SJ, Booth IR (November 2010). "Mechanism of ligand-gated potassium efflux in bacterial pathogens". Proceedings of the National Academy of Sciences of the United States of America. 107 (46): 19784–9. Bibcode:2010PNAS..10719784R. doi: 10.1073/pnas.1012716107 . PMC 2993342 . PMID 21041667.
- ↑ Nakamura C, Kikuchi T, Burgess JG, Matsunaga T (July 1995). "Iron-regulated expression and membrane localization of the magA protein in Magnetospirillum sp. strain AMB-1". Journal of Biochemistry. 118 (1): 23–7. doi:10.1093/oxfordjournals.jbchem.a124884. PMID 8537318.
- ↑ Uebe R, Henn V, Schüler D (March 2012). "The MagA protein of Magnetospirilla is not involved in bacterial magnetite biomineralization". Journal of Bacteriology. 194 (5): 1018–23. doi:10.1128/JB.06356-11. PMC 3294778 . PMID 22194451.
- ↑ Southworth TW, Guffanti AA, Moir A, Krulwich TA (October 2001). "GerN, an endospore germination protein of Bacillus cereus, is an Na(+)/H(+)-K(+) antiporter". Journal of Bacteriology. 183 (20): 5896–903. doi:10.1128/JB.183.20.5896-5903.2001. PMC 99667 . PMID 11566988.
- ↑ Tani K, Watanabe T, Matsuda H, Nasu M, Kondo M (1996-01-01). "Cloning and sequencing of the spore germination gene of Bacillus megaterium ATCC 12872: similarities to the NaH-antiporter gene of Enterococcus hirae". Microbiology and Immunology. 40 (2): 99–105. doi: 10.1111/j.1348-0421.1996.tb03323.x . PMID 8867604. S2CID 7130059.