
A mitochondrion is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term mitochondrion was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name.
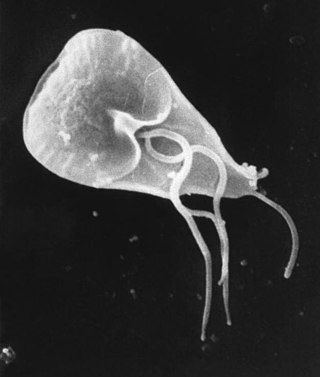
Giardia is a genus of anaerobic flagellated protozoan parasites of the phylum Metamonada that colonise and reproduce in the small intestines of several vertebrates, causing the disease giardiasis. Their life cycle alternates between a swimming trophozoite and an infective, resistant cyst. Giardia were first described by the Dutch microscopist Antonie van Leeuwenhoek in 1681. The genus is named after French zoologist Alfred Mathieu Giard.

Excavata is an extensive and diverse but paraphyletic group of unicellular Eukaryota. The group was first suggested by Simpson and Patterson in 1999 and the name latinized and assigned a rank by Thomas Cavalier-Smith in 2002. It contains a variety of free-living and symbiotic protists, and includes some important parasites of humans such as Giardia and Trichomonas. Excavates were formerly considered to be included in the now obsolete Protista kingdom. They were distinguished from other lineages based on electron-microscopic information about how the cells are arranged. They are considered to be a basal flagellate lineage.

The parabasalids are a group of flagellated protists within the supergroup Excavata. Most of these eukaryotic organisms form a symbiotic relationship in animals. These include a variety of forms found in the intestines of termites and cockroaches, many of which have symbiotic bacteria that help them digest cellulose in woody plants. Other species within this supergroup are known parasites, and include human pathogens.
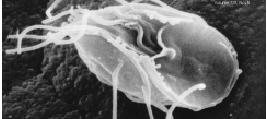
The metamonads are a large group of flagellate amitochondriate microscopic eukaryotes. Their composition is not entirely settled, but they include the retortamonads, diplomonads, and possibly the parabasalids and oxymonads as well. These four groups are all anaerobic, occurring mostly as symbiotes or parasites of animals, as is the case with Giardia lamblia which causes diarrhea in mammals.

A hydrogenosome is a membrane-enclosed organelle found in some anaerobic ciliates, flagellates, and fungi. Hydrogenosomes are highly variable organelles that have presumably evolved from protomitochondria to produce molecular hydrogen and ATP in anaerobic conditions.
The Oxymonads are a group of flagellated protists found exclusively in the intestines of animals, mostly termites and other wood-eating insects. Along with the similar parabasalid flagellates, they harbor the symbiotic bacteria that are responsible for breaking down cellulose. There is no evidence for presence of mitochondria in oxymonads and 3 species have been shown to completely lack any molecular markers of mitochondria.
A symbiotic eukaryote that lives in the hindgut of termites, Streblomastix is a protist associated with a community of ectosymbiotic bacteria.

Breviata anathema is a single-celled flagellate amoeboid eukaryote, previously studied under the name Mastigamoeba invertens. The cell lacks mitochondria, much like the pelobionts to which the species was previously assigned, but has remnant mitochondrial genes, and possesses an organelle believed to be a modified anaerobic mitochondrion, similar to the mitosomes and hydrogenosomes found in other eukaryotes that live in low-oxygen environments.
Trimastix is a genus of excavate protists, the sole occupant of the order Trimastigida. Trimastix are bacterivorous, free living and anaerobic. It was first observed in 1881 by William Kent. There are few known species, and the genus's role in the ecosystem is largely unknown. However, it is known that they generally live in marine environments within the tissues of decaying organisms to maintain an anoxic environment. Much interest in this group is related to its close association with other members of Preaxostyla. These organisms do not have classical mitochondria, and as such, much of the research involving these microbes is aimed at investigating the evolution of mitochondria.
Anaeromonadea, also known as Preaxostyla, is a class of excavate protists, comprising the oxymonads, Trimastix, and Paratrimastix. This group is studied as a model system for reductive evolution of mitochondria, because it includes both organisms with anaerobic mitochondrion-like organelles, and those that have completely lost their mitochondria.
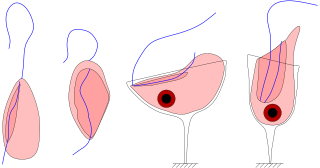
Jakobids are an order of free-living, heterotrophic, flagellar eukaryotes in the supergroup Excavata. They are small, and can be found in aerobic and anaerobic environments. The order Jakobida, believed to be monophyletic, consists of only twenty species at present, and was classified as a group in 1993. There is ongoing research into the mitochondrial genomes of jakobids, which are unusually large and bacteria-like, evidence that jakobids may be important to the evolutionary history of eukaryotes.

Proteromonas is a genus of single-celled biflagellated microbial eukaryotes belonging to the Superphylum Stramenopiles which are characterized by the presence of tripartite, hair-like structures on the anteriorly-directed larger of the two flagella. Proteromonas on the other hand are notable by having tripartite hairs called somatonemes not on the flagella but on the posterior of the cell. Proteromonas are closely related to Karotomorpha and Blastocystis, which belong to the Opalines group.

Mastigamoeba is a genus of pelobionts, and treated by some as members of the Archamoebae group of protists. Mastigamoeba are characterized as anaerobic, amitochondriate organisms that are polymorphic. Their dominant life cycle stage is as an amoeboid flagellate. Species are typically free living, though endobiotic species have been described.
Monocercomonoides is a genus of flagellate Excavata belonging to the order Oxymonadida. It was established by Bernard V. Travis and was first described as those with "polymastiginid flagellates having three anterior flagella and a trailing one originating at a single basal granule located in front of the anteriorly positioned nucleus, and a more or less well-defined axostyle". It is the first eukaryotic genus to be found to completely lack mitochondria, and all hallmark proteins responsible for mitochondrial function. The genus also lacks any other mitochondria related organelles (MROs) such as hydrogenosomes or mitosomes. Data suggests that the absence of mitochondria is not an ancestral feature, but rather due to secondary loss. Monocercomonoides sp. was found to obtain energy through an enzymatic action of nutrients absorbed from the environment. The genus has replaced the iron-sulfur cluster assembly pathway with a cytosolic sulfur mobilization system, likely acquired by horizontal gene transfer from a eubacterium of a common ancestor of oxymonads. These organisms are significant because they undermine assumptions that eukaryotes must have mitochondria to properly function. The genome of Monocercomonoides exilis has approximately 82 million base pairs, with 18 152 predicted protein-coding genes.

Andrew J. Roger is a Canadian-Australian molecular biologist and evolutionary bioinformatician. He is currently a professor in the Department of Biochemistry and Molecular Biology at Dalhousie University and was the founding director of the inter-departmental Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB).
Stygiella /ˌstɪ.d͡ʒiˈɛ.lə/ is a genus of free-living marine flagellates belonging to the family Stygiellidae in the jakobids (excavata).
Anaeramoeba is a genus of anaerobic protists of uncertain phylogenetic position, first described in 2016.

Stygiellidae is a family of free-living marine flagellates belonging to the order Jakobida, a deep-branching lineage within the eukaryotic supergroup Discoba. They are unicellular organisms that commonly inhabit anoxic, sulfide-rich and ammonium-rich marine habitats worldwide.
Paratrimastix is a genus of free-living freshwater anaerobic excavate protists from the group Metamonada, that was segregated from the genus Trimastix in 2015. The best studied species is Paratrimastix pyriformis.











