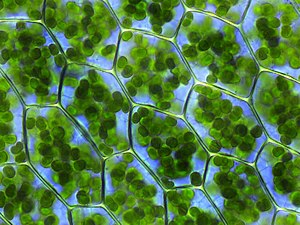
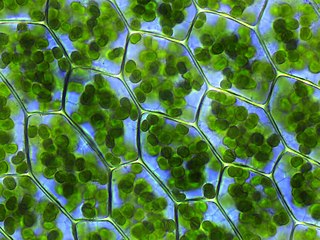
A chloroplast is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it in the energy-storage molecules ATP and NADPH while freeing oxygen from water in the cells. The ATP and NADPH is then used to make organic molecules from carbon dioxide in a process known as the Calvin cycle. Chloroplasts carry out a number of other functions, including fatty acid synthesis, amino acid synthesis, and the immune response in plants. The number of chloroplasts per cell varies from one, in unicellular algae, up to 100 in plants like Arabidopsis and wheat.
Microevolution is the change in allele frequencies that occurs over time within a population. This change is due to four different processes: mutation, selection, gene flow and genetic drift. This change happens over a relatively short amount of time compared to the changes termed macroevolution.
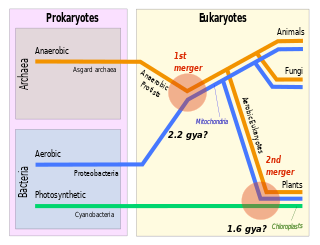
Symbiogenesis is the leading evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms. The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells are descended from formerly free-living prokaryotes taken one inside the other in endosymbiosis. Mitochondria appear to be phylogenetically related to Rickettsiales bacteria, while chloroplasts are thought to be related to cyanobacteria.

Polyploidy is a condition in which the cells of an organism have more than one pair of (homologous) chromosomes. Most species whose cells have nuclei (eukaryotes) are diploid, meaning they have two complete sets of chromosomes, one from each of two parents; each set contains the same number of chromosomes, and the chromosomes are joined in pairs of homologous chromosomes. However, some organisms are polyploid. Polyploidy is especially common in plants. Most eukaryotes have diploid somatic cells, but produce haploid gametes by meiosis. A monoploid has only one set of chromosomes, and the term is usually only applied to cells or organisms that are normally diploid. Males of bees and other Hymenoptera, for example, are monoploid. Unlike animals, plants and multicellular algae have life cycles with two alternating multicellular generations. The gametophyte generation is haploid, and produces gametes by mitosis; the sporophyte generation is diploid and produces spores by meiosis.
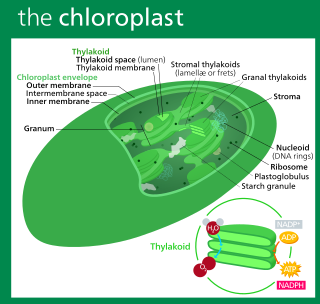
The plastid is a membrane-bound organelle found in the cells of plants, algae, and some other eukaryotic organisms. They are considered to be intracellular endosymbiotic cyanobacteria. Examples include chloroplasts, chromoplasts, and leucoplasts.
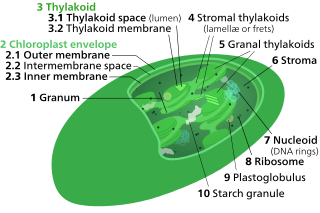
Thylakoids are membrane-bound compartments inside chloroplasts and cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thylakoids frequently form stacks of disks referred to as grana. Grana are connected by intergranal or stromal thylakoids, which join granum stacks together as a single functional compartment.
Endoreduplication is replication of the nuclear genome in the absence of mitosis, which leads to elevated nuclear gene content and polyploidy. Endoreplication can be understood simply as a variant form of the mitotic cell cycle (G1-S-G2-M) in which mitosis is circumvented entirely, due to modulation of cyclin-dependent kinase (CDK) activity. Examples of endoreplication characterized in arthropod, mammalian, and plant species suggest that it is a universal developmental mechanism responsible for the differentiation and morphogenesis of cell types that fulfill an array of biological functions. While endoreplication is often limited to specific cell types in animals, it is considerably more widespread in plants, such that polyploidy can be detected in the majority of plant tissues.

Paleopolyploidy is the result of genome duplications which occurred at least several million years ago (MYA). Such an event could either double the genome of a single species (autopolyploidy) or combine those of two species (allopolyploidy). Because of functional redundancy, genes are rapidly silenced or lost from the duplicated genomes. Most paleopolyploids, through evolutionary time, have lost their polyploid status through a process called diploidization, and are currently considered diploids, e.g., baker's yeast, Arabidopsis thaliana, and perhaps humans.

The Archaeplastida are a major group of eukaryotes, comprising the photoautotrophic red algae (Rhodophyta), green algae, land plants, and the minor group glaucophytes. It also includes the non-photosynthetic lineage Rhodelphidia, a predatorial (eukaryotrophic) flagellate that is sister to the Rhodophyta, and probably the microscopic picozoans. The Archaeplastida have chloroplasts that are surrounded by two membranes, suggesting that they were acquired directly through a single endosymbiosis event by feeding on a cyanobacterium. All other groups which have chloroplasts, besides the amoeboid genus Paulinella, have chloroplasts surrounded by three or four membranes, suggesting they were acquired secondarily from red or green algae. Unlike red and green algae, glaucophytes have never been involved in secondary endosymbiosis events.
Evolutionary developmental biology (evo-devo) is the study of developmental programs and patterns from an evolutionary perspective. It seeks to understand the various influences shaping the form and nature of life on the planet. Evo-devo arose as a separate branch of science rather recently. An early sign of this occurred in 1999.

Paulinella is a genus of at least eleven species including both freshwater and marine amoeboids. Like many members of euglyphids it is covered by rows of siliceous scales, and use filose pseudopods to crawl over the substrate of the benthic zone.
The CoRR hypothesis states that the location of genetic information in cytoplasmic organelles permits regulation of its expression by the reduction-oxidation ("redox") state of its gene products.

Guillardia is a genus of marine biflagellate cryptomonad algae with a plastid obtained through secondary endosymbiosis of a red alga.
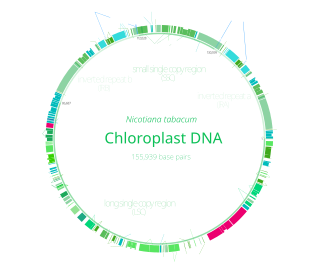
Chloroplast DNA (cpDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, a great number of chloroplast DNAs from various species have been sequenced.

Leslie David Gottlieb (1936–2012) was a United States biologist described by the Botanical Society of America as "one of the most influential plant evolutionary biologists over the past several decades". He was employed at the University of California, Davis for 34 years, and published widely. In addition to his primary work in plant genetics, Gottlieb was an advocate for rare and endangered plant conservation.
The evolution of photosynthesis refers to the origin and subsequent evolution of photosynthesis, the process by which light energy is used to assemble sugars from carbon dioxide and a hydrogen and electron source such as water. The process of photosynthesis was discovered by Jan Ingenhousz, a Dutch-born British physician and scientist, first publishing about it in 1779.
In Embryology a phylotypic stage or phylotypic period is a particular developmental stage or developmental period during mid-embryogenesis where embryos of related species within a phylum express the highest degree of morphological and molecular resemblance. Recent molecular studies in various plant and animal species were able to quantify the expression of genes covering crucial stages of embryo development and found that during the morphologically defined phylotypic period the evolutionary oldest genes, genes with similar temporal expression patterns, and genes under strongest purifying selection are most active throughout the phylotypic period.
Diploidization is the process of converting a polyploid genome back into a diploid one. Polyploidy is a product of whole genome duplication (WGD) and is followed by diploidization as a result of genome shock. The plant kingdom has undergone multiple events of polyploidization followed by diploidization in both ancient and recent lineages. It has also been hypothesized that vertebrate genomes have gone through two rounds of paleopolyploidy. The mechanisms of diploidization are poorly understood but patterns of chromosomal loss and evolution of novel genes are observed in the process.
A plastid is a membrane-bound organelle found in plants, algae and other eukaryotic organisms that contribute to the production of pigment molecules. Most plastids are photosynthetic, thus leading to color production and energy storage or production. There are many types of plastids in plants alone, but all plastids can be separated based on the number of times they have undergone endosymbiotic events. Currently there are three types of plastids; primary, secondary and tertiary. Endosymbiosis is reputed to have led to the evolution of eukaryotic organisms today, although the timeline is highly debated.