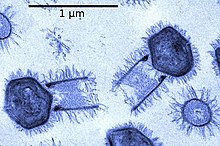
Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas, and cirrhosis. It is the sole accepted family in the order Blubervirales.

Mimivirus is a genus of giant viruses, in the family Mimiviridae. Amoeba serve as their natural hosts. This genus contains a single identified species named Acanthamoeba polyphaga mimivirus (APMV). It also refers to a group of phylogenetically related large viruses.
Metaviridae is a family of viruses which exist as Ty3-gypsy LTR retrotransposons in a eukaryotic host's genome. They are closely related to retroviruses: members of the family Metaviridae share many genomic elements with retroviruses, including length, organization, and genes themselves. This includes genes that encode reverse transcriptase, integrase, and capsid proteins. The reverse transcriptase and integrase proteins are needed for the retrotransposon activity of the virus. In some cases, virus-like particles can be formed from capsid proteins.
Pseudoviridae is a family of viruses, which includes three genera.

A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898, more than 11,000 of the millions of virus species have been described in detail. The study of viruses is known as virology, a subspeciality of microbiology.

Mimivirus-dependent virus Sputnik is a subviral agent that reproduces in amoeba cells that are already infected by a certain helper virus; Sputnik uses the helper virus's machinery for reproduction and inhibits replication of the helper virus. It is known as a virophage, in analogy to the term bacteriophage.

Mimiviridae is a family of viruses. Amoeba and other protists serve as natural hosts. The family is divided in up to 4 subfamilies. Viruses in this family belong to the nucleocytoplasmic large DNA virus clade (NCLDV), also referred to as giant viruses.

Halspiviridae is a family of viruses that consists of a single genus, Salterprovirus, which consists of a single recognised species; Salterprovirus His1. This virus was isolated from hypersaline water in Australia and was able to be cultured on the halophilic archaeon Haloarcula hispanica. Like many other archaeoviruses, His1 has an approximately limoniform (lemon-shaped) virion.
Mamavirus is a large and complex virus in the Group I family Mimiviridae. The virus is exceptionally large, and larger than many bacteria. Mamavirus and other mimiviridae belong to nucleocytoplasmic large DNA virus (NCLDVs) family. Mamavirus can be compared to the similar complex virus mimivirus; mamavirus was so named because it is similar to but larger than mimivirus.

Vesivirus is a genus of viruses, in the family Caliciviridae. Swine, sea mammals, and felines serve as natural hosts. There are two species in this genus. Diseases associated with this genus include: respiratory disease, Feline calicivirus (FCV); conjunctivitis, and respiratory disease.
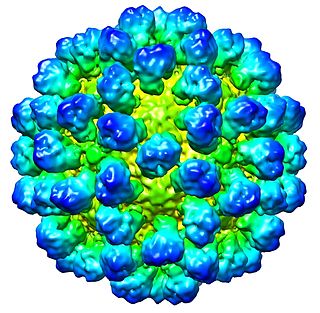
Lagovirus is a genus of viruses, in the family Caliciviridae. Lagomorphs serve as natural hosts. There are two species in this genus. Diseases associated with this genus include: necrotizing hepatitis leading to fatal hemorrhages.

Cafeteria roenbergensis virus (CroV) is a giant virus that infects the marine bicosoecid flagellate Cafeteria roenbergensis, a member of the microzooplankton community.
A giant virus, sometimes referred to as a girus, is a very large virus, some of which are larger than typical bacteria. All known giant viruses belong to the phylum Nucleocytoviricota.

Megavirus is a viral genus, phylogenetically related to Acanthamoeba polyphaga mimivirus (APMV). In colloquial speech, Megavirus chilense is more commonly referred to as just "Megavirus". Until the discovery of pandoraviruses in 2013, it had the largest capsid diameter of all known viruses, as well as the largest and most complex genome among all known viruses.
Mason-Pfizer monkey virus (M-PMV), formerly Simian retrovirus (SRV), is a species of retroviruses that usually infect and cause a fatal immune deficiency in Asian macaques. The ssRNA virus appears sporadically in mammary carcinoma of captive macaques at breeding facilities which expected as the natural host, but the prevalence of this virus in feral macaques remains unknown. M-PMV was transmitted naturally by virus-containing body fluids, via biting, scratching, grooming, and fighting. Cross contaminated instruments or equipment (fomite) can also spread this virus among animals.
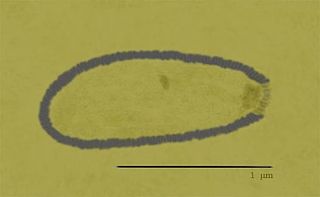
Alphapithovirus, is a genus of giant virus known from two species, Alphapithovirus sibericum, which infects amoebas, and Alphapithovirus massiliense. It is DNA-based and is a member of the nucleocytoplasmic large DNA viruses clade. It was discovered in 2014, when a viable specimen was found in a 30,000-year-old ice core harvested from permafrost in Siberia, Russia.

Mimivirus-dependent virus Zamilon, or Zamilon, is a virophage, a group of small DNA viruses that infect protists and require a helper virus to replicate; they are a type of satellite virus. Discovered in 2013 in Tunisia, infecting Acanthamoeba polyphaga amoebae, Zamilon most closely resembles Sputnik, the first virophage to be discovered. The name is Arabic for "the neighbour". Its spherical particle is 50–60 nm in diameter, and contains a circular double-stranded DNA genome of around 17 kb, which is predicted to encode 20 polypeptides. A related strain, Zamilon 2, has been identified in North America.

Faustovirus is a genus of giant virus which infects amoebae associated with humans. The virus was first isolated in 2015 and shown to be around 0.2 micrometers in diameter with a double stranded DNA genome of 466 kilobases predicted to encode 451 proteins. Although classified as a nucleocytoplasmic large DNA virus (NCLDV), faustoviruses share less than a quarter of their genes with other NCLDVs; however, ~46% are homologous to bacterial genes and the remainder are orphan genes (ORFans). Specifically, the gene encoding the major capsid protein (MCP) of faustovirus is different than that of its most closely related giant virus, asfivirus, as well as other NCLDVs. In asfivirus, the gene encoding MCP is a single genomic fragment of ~2000 base pairs (bp), however, in faustovirus the MCP is encoded by 13 exons separated by 12 large introns. The exons have a mean length of 149 bp and the introns have a mean length of 1,273 bp. The presence of introns in faustovirus genes is highly unusual for viruses.
Nucleocytoviricota is a phylum of viruses. Members of the phylum are also known as the nucleocytoplasmic large DNA viruses (NCLDV), which serves as the basis of the name of the phylum with the suffix -viricota for virus phylum. These viruses are referred to as nucleocytoplasmic because they are often able to replicate in both the host's cell nucleus and cytoplasm.