
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part of biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.

Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology.

A ρ factor is a bacterial protein involved in the termination of transcription. Rho factor binds to the transcription terminator pause site, an exposed region of single stranded RNA after the open reading frame at C-rich/G-poor sequences that lack obvious secondary structure.
A sigma factor is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP).
A transcriptional activator is a protein that increases transcription of a gene or set of genes. Activators are considered to have positive control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain".

The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a typical prokaryote is circular, and its length is very large compared to the cell dimensions, so it needs to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.

DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
In molecular biology, a termination factor is a protein that mediates the termination of RNA transcription by recognizing a transcription terminator and causing the release of the newly made mRNA. This is part of the process that regulates the transcription of RNA to preserve gene expression integrity and are present in both eukaryotes and prokaryotes, although the process in bacteria is more widely understood. The most extensively studied and detailed transcriptional termination factor is the Rho (ρ) protein of E. coli.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The total result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
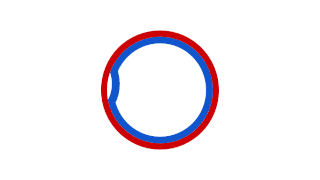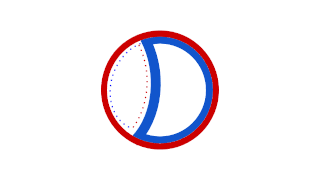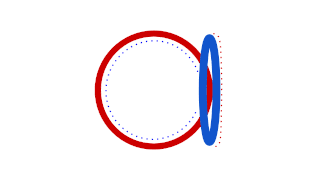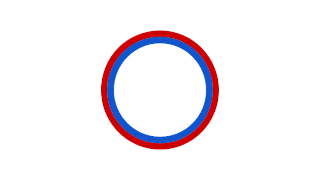
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.

fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.

The bacterial one-hybrid (B1H) system is a method for identifying the sequence-specific target site of a DNA-binding domain. In this system, a given transcription factor (TF) is expressed as a fusion to a subunit of RNA polymerase. In parallel, a library of randomized oligonucleotides representing potential TF target sequences are cloned into a separate vector containing the selectable genes HIS3 and URA3. If the DNA-binding domain (bait) binds a potential DNA target site (prey) in vivo, it will recruit RNA polymerase to the promoter and activate transcription of the reporter genes in that clone. The two reporter genes, HIS3 and URA3, allow for positive and negative selections, respectively. At the end of the process, positive clones are sequenced and examined with motif-finding tools in order to resolve the favoured DNA target sequence.
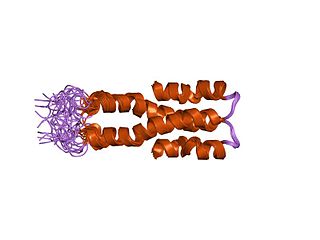
Histone-like nucleoid-structuring protein (H-NS), is one of twelve nucleoid-associated proteins (NAPs) whose main function is the organization of genetic material, including the regulation of gene expression via xenogeneic silencing. H-NS is characterized by an N-terminal domain (NTD) consisting of two dimerization sites, a linker region that is unstructured and a C-terminal domain (CTD) that is responsible for DNA-binding. Though it is a small protein, it provides essential nucleoid compaction and regulation of genes and is highly expressed, functioning as a dimer or multimer. Change in temperature causes H-NS to be dissociated from the DNA duplex, allowing for transcription by RNA polymerase, and in specific regions lead to pathogenic cascades in enterobacteria such as Escherichia coli and the four Shigella species.
The locus of enterocyte effacement-encoded regulator (Ler) is a regulatory protein that controls bacterial pathogenicity of enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic Escherichia coli (EHEC). More specifically, Ler regulates the locus of enterocyte effacement (LEE) pathogenicity island genes, which are responsible for creating intestinal attachment and effacing lesions and subsequent diarrhea: LEE1, LEE2, and LEE3. LEE1, 2, and 3 carry the information necessary for a type III secretion system. The transcript encoding the Ler protein is the open reading frame 1 on the LEE1 operon.














