Developmental biology is the study of the process by which animals and plants grow and develop. Developmental biology also encompasses the biology of regeneration, asexual reproduction, metamorphosis, and the growth and differentiation of stem cells in the adult organism.

An embryo is the initial stage of development for a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm cell. The resulting fusion of these two cells produces a single-celled zygote that undergoes many cell divisions that produce cells known as blastomeres. The blastomeres are arranged as a solid ball that when reaching a certain size, called a morula, takes in fluid to create a cavity called a blastocoel. The structure is then termed a blastula, or a blastocyst in mammals.

Caenorhabditis elegans is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek caeno- (recent), rhabditis (rod-like) and Latin elegans (elegant). In 1900, Maupas initially named it Rhabditides elegans. Osche placed it in the subgenus Caenorhabditis in 1952, and in 1955, Dougherty raised Caenorhabditis to the status of genus.
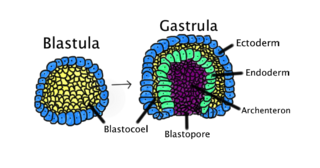
Gastrulation is the stage in the early embryonic development of most animals, during which the blastula, or in mammals the blastocyst, is reorganized into a two-layered or three-layered embryo known as the gastrula. Before gastrulation, the embryo is a continuous epithelial sheet of cells; by the end of gastrulation, the embryo has begun differentiation to establish distinct cell lineages, set up the basic axes of the body, and internalized one or more cell types including the prospective gut.

The blastocyst is a structure formed in the early embryonic development of mammals. It possesses an inner cell mass (ICM) also known as the embryoblast which subsequently forms the embryo, and an outer layer of trophoblast cells called the trophectoderm. This layer surrounds the inner cell mass and a fluid-filled cavity known as the blastocoel. In the late blastocyst, the trophectoderm is known as the trophoblast. The trophoblast gives rise to the chorion and amnion, the two fetal membranes that surround the embryo. The placenta derives from the embryonic chorion and the underlying uterine tissue of the mother.

Howard Robert Horvitz ForMemRS NAS AAA&S APS NAM is an American biologist best known for his research on the nematode worm Caenorhabditis elegans, for which he was awarded the 2002 Nobel Prize in Physiology or Medicine, together with Sydney Brenner and John E. Sulston, whose "seminal discoveries concerning the genetic regulation of organ development and programmed cell death" were "important for medical research and have shed new light on the pathogenesis of many diseases".

Sir John Edward Sulston was a British biologist and academic who won the Nobel Prize in Physiology or Medicine for his work on the cell lineage and genome of the worm Caenorhabditis elegans in 2002 with his colleagues Sydney Brenner and Robert Horvitz at the MRC Laboratory of Molecular Biology. He was a leader in human genome research and Chair of the Institute for Science, Ethics and Innovation at the University of Manchester. Sulston was in favour of science in the public interest, such as free public access of scientific information and against the patenting of genes and the privatisation of genetic technologies.
A germ layer is a primary layer of cells that forms during embryonic development. The three germ layers in vertebrates are particularly pronounced; however, all eumetazoans produce two or three primary germ layers. Some animals, like cnidarians, produce two germ layers making them diploblastic. Other animals such as bilaterians produce a third layer between these two layers, making them triploblastic. Germ layers eventually give rise to all of an animal's tissues and organs through the process of organogenesis.
Organogenesis is the phase of embryonic development that starts at the end of gastrulation and continues until birth. During organogenesis, the three germ layers formed from gastrulation form the internal organs of the organism.
Cre-Lox recombination is a site-specific recombinase technology, used to carry out deletions, insertions, translocations and inversions at specific sites in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type or be triggered by a specific external stimulus. It is implemented both in eukaryotic and prokaryotic systems. The Cre-lox recombination system has been particularly useful to help neuroscientists to study the brain in which complex cell types and neural circuits come together to generate cognition and behaviors. NIH Blueprint for Neuroscience Research has created several hundreds of Cre driver mouse lines which are currently used by the worldwide neuroscience community.

In genetics, Flp-FRT recombination is a site-directed recombination technology, increasingly used to manipulate an organism's DNA under controlled conditions in vivo. It is analogous to Cre-lox recombination but involves the recombination of sequences between short flippase recognition target (FRT) sites by the recombinase flippase (Flp) derived from the 2 µ plasmid of baker's yeast Saccharomyces cerevisiae.
An equivalence group is a set of unspecified cells that have the same developmental potential or ability to adopt various fates. Our current understanding suggests that equivalence groups are limited to cells of the same ancestry, also known as sibling cells. Often, cells of an equivalence group adopt different fates from one another.
Within the field of developmental biology, one goal is to understand how a particular cell develops into a final cell type, known as fate determination. Within an embryo, several processes play out at the cellular and tissue level to create an organism. These processes include cell proliferation, differentiation, cellular movement and programmed cell death. Each cell in an embryo receives molecular signals from neighboring cells in the form of proteins, RNAs and even surface interactions. Almost all animals undergo a similar sequence of events during very early development, a conserved process known as embryogenesis. During embryogenesis, cells exist in three germ layers, and undergo gastrulation. While embryogenesis has been studied for more than a century, it was only recently that scientists discovered that a basic set of the same proteins and mRNAs are involved in embryogenesis. Evolutionary conservation is one of the reasons that model systems such as the fly, the mouse, and other organisms are used as models to study embryogenesis and developmental biology. Studying model organisms provides information relevant to other animals, including humans. While studying the different model systems, cells fate was discovered to be determined via multiple ways, two of which are by the combination of transcription factors the cells have and by the cell-cell interaction. Cells' fate determination mechanisms were categorized into three different types, autonomously specified cells, conditionally specified cells, or syncytial specified cells. Furthermore, the cells' fate was determined mainly using two types of experiments, cell ablation and transplantation. The results obtained from these experiments, helped in identifying the fate of the examined cells.

Janet Rossant, is a developmental biologist well known for her contributions to the understanding of the role of genes in embryo development. She is a world renowned leader in developmental biology. Her current research interests focus on stem cells, molecular genetics, and developmental biology. Specifically, she uses cellular and genetic manipulation techniques to study how genes control both normal and abnormal development of early mouse embryos. Rossant has discovered information on embryo development, how multiple types of stem cells are established, and the mechanisms by which genes control development. In 1998, her work helped lead to the discovery of the trophoblast stem cell, which has assisted in showing how congenital anomalies in the heart, blood vessels, and placenta can occur.

Rosa Susan Penelope Beddington FRS was a British biologist whose career had a major impact on developmental biology.

Elizabeth Jane Robertson is a British developmental biologist based at the Sir William Dunn School of Pathology, University of Oxford. She is Professor of Developmental Biology at Oxford and a Wellcome Trust Principal Research Fellow. She is best known for her pioneering work in developmental genetics, showing that genetic mutations could be introduced into the mouse germ line by using genetically altered embryonic stem cells. This discovery opened up a major field of experimentation for biologists and clinicians.

Cell lineage denotes the developmental history of a tissue or organ from the fertilized embryo. This is based on the tracking of an organism's cellular ancestry due to the cell divisions and relocation as time progresses, this starts with the originator cells and finishing with a mature cell that can no longer divide.
The Spemann-Mangold organizer is a group of cells that are responsible for the induction of the neural tissues during development in amphibian embryos. First described in 1924 by Hans Spemann and Hilde Mangold, the introduction of the organizer provided evidence that the fate of cells can be influenced by factors from other cell populations. This discovery significantly impacted the world of developmental biology and fundamentally changed the understanding of early development.

The dorsal lip of the blastopore is a structure that forms during early embryonic development and is important for its role in organizing the germ layers. The dorsal lip is formed during early gastrulation as folding of tissue along the involuting marginal zone of the blastocoel forms an opening known as the blastopore. It is particularly important for its role in neural induction through the default model, where signaling from the dorsal lip protects a region of the epiblast from becoming epidermis, thus allowing it to develop to its default neural tissue.

Genome editing of synthetic target arrays for lineage tracing (GESTALT) is a method used to determine the developmental lineages of cells in multicellular systems. GESTALT involves introducing a small DNA barcode that contains regularly spaced CRISPR/Cas9 target sites into the genomes of progenitor cells. Alongside the barcode, Cas9 and sgRNA are introduced into the cells. Mutations in the barcode accumulate during the course of cell divisions and the unique combination of mutations in a cell's barcode can be determined by DNA or RNA sequencing to link it to a developmental lineage.














