
Genetic studies show that Russians are closest to Poles, Belarusians, Ukrainians and to other Slavs as well as to Estonians, Latvians, Lithuanians, Hungarians. [1]

Genetic studies show that Russians are closest to Poles, Belarusians, Ukrainians and to other Slavs as well as to Estonians, Latvians, Lithuanians, Hungarians. [1]
Eight Y chromosome haplogroup subclades, including R1a, N3, I1b, R1b, I2a, J2, N2, and E3b all together, account for >95% of the total Russian Y chromosomal pool. Of the 1228 samples, 11/1228 (0.9%) were classified up to the root level of haplogroups F and K. Only 9/1228 samples (0.7%) fell into haplogroups C, Q, and R2 which are specific to East and South Asian populations. [1]
The top four Y-DNA haplogroups among the sample of 1228 Russians are: [1]
Eight Y chromosome haplogroup subclades, including R1a, N3, I1b, R1b, I1a, J2, N2, and E3b all together, account for >95% of the total Russian Y chromosomal pool. Of the 1228 samples, 11/1228 (0.9%) were classified up to the root level of haplogroups F and K. Only 9/1228 samples (0.7%) fell into haplogroups C, Q, and R2 which are specific to East and South Asian populations. [1]
The mitochondrial gene pool of Russians are represented by mtDNA types belonging to typical West Eurasian groups. East Eurasian admixture was shown to be minimal and existed in low frequencies in the form of Haplogroup M. [2] [3] The same studies indicate Eurasian haplogroups present at a frequency of 97.8% and 98.5% among a sample of 325 and 201 Russians respectively. [2] [3]

Autosomally, Russians are most similar to populations in Eastern Europe followed by other Eurasian groups. [5] Genetic research suggests higher amounts of Siberian admixture among Northern Russians than Central and Southern Russians. Eastern Siberian-like ancestry was found at an average frequency of ~12% among Northern Russians. This Eastern Siberian-like ancestry is maximized among modern Nganasan people and a Bronze Age specimen from Southern Siberia (Krasnoyarsk_Krai_BA), suggesting the assimilation and slavification of Uralic ethnic groups during the expansion of early Russians. Other Russians carried it at a lower frequency of around 4% similarly to other Slavic populations. [6] [7] [8] A study by Wang et al. argued that the levels of "Eastern Siberian" ancestry among Russians, but also Finns, may be linked to the diffusion of paternal haplogroup N-M231. [9]
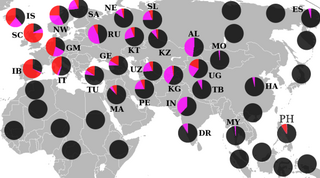
A large full-genome study by Triska et al. 2017 on Russians and other ethnic groups, stretching from the Baltic region to Lake Baikal, found that Russians are closely related to other Slavic Peoples, followed by other European populations. According to the Russian researchers, "Ukrainians, Belarusians and Russians have almost identical proportions of Caucasus and Northern European components and have virtually no Asian influence", contradicting the previous studies mentioned. However, many ethnic minority groups from Russia were found to be characterized by the presence of significant higher amounts of Asian components, yet European ancestry also makes up the majority of their respective gene pool: "European components account for 50% - 90% of admixture vectors in both Turkic and Uralic speakers of the Volga-Ural region". Other ethnic minorities, such as from the Caucasus region, harbored predominantly "Caucasus and Iranian" components. [10]
A genome-wide sequence analysis on the population of Russia by Zhernakova et al. 2020, found that Russia as a whole, displays significant heterogeneity. Ethnic Russians primarily descended from the early Slavic peoples, which diverged from other Indo-Europeans, and expanded from Eastern Europe eastwards. Subsequently Russians expanded further eastwards, later coming into contact with various other groups, such as Uralic, Turkic, Iranian, Mongolic and Tungusic peoples as well as Paleo-Siberian groups of Siberia. Geneflow between Asian minority groups and Russians contributed to the overall pattern of genome diversity across the different ethno-linguistic groups of Russia. [11] [12]
Genetics and archaeogenetics of South Asia is the study of the genetics and archaeogenetics of the ethnic groups of South Asia. It aims at uncovering these groups' genetic histories. The geographic position of the Indian subcontinent makes its biodiversity important for the study of the early dispersal of anatomically modern humans across Asia.
Haplogroup R1, or R-M173, is a Y-chromosome DNA haplogroup. A primary subclade of Haplogroup R (R-M207), it is defined by the SNP M173. The other primary subclade of Haplogroup R is Haplogroup R2 (R-M479).
Haplogroup R, or R-M207, is a Y-chromosome DNA haplogroup. It is both numerous and widespread amongst modern populations.

Tungusic peoples are an ethnolinguistic group formed by the speakers of Tungusic languages. They are native to Siberia, China, and Mongolia.
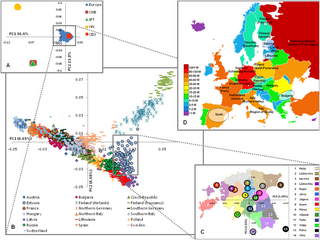
The genetic history of Europe includes information around the formation, ethnogenesis, and other DNA-specific information about populations indigenous, or living in Europe.
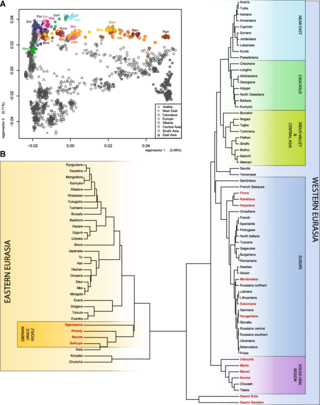
Genetic studies on Sami is the genetic research that have been carried out on the Sami people. The Sami languages belong to the Uralic languages family of Eurasia.

Haplogroup R1a, or haplogroup R-M420, is a human Y-chromosome DNA haplogroup which is distributed in a large region in Eurasia, extending from Scandinavia and Central Europe to Central Asia, southern Siberia and South Asia.

Haplogroup R1b (R-M343), previously known as Hg1 and Eu18, is a human Y-chromosome haplogroup.
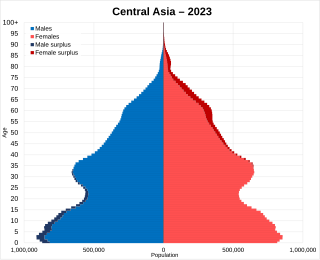
The nations which make up Central Asia are five of the former Soviet republics: Kazakhstan, Kyrgyzstan, Turkmenistan, Tajikistan and Uzbekistan, which have a total population of about 76 million. Afghanistan is not always considered part of the region, but when it is, Central Asia has a total population of about 122 million (2016); Mongolia and Xinjiang is also sometimes considered part of Central Asia due to its Central Asian cultural ties and traditions, although geographically it is East Asian. Most central Asians belong to religions which were introduced to the area within the last 1,500 years, such as Sunni Islam, Shia Islam, Ismaili Islam, Tengriism and Syriac Christianity. Buddhism, however, was introduced to Central Asia over 2,200 years ago, and Zoroastrianism, over 2,500 years ago.
The genetic history of the Indigenous peoples of the Americas is divided into two distinct periods: the initial peopling of the Americas during about 20,000 to 14,000 years ago, and European contact, after about 500 years ago. The first period of the genetic history of Indigenous Americans is the determinant factor for the number of genetic lineages, zygosity mutations, and founding haplotypes present in today's Indigenous American populations.
Population genetics research has been conducted on the ancestry of the modern Turkish people in Turkey. Such studies are relevant for the demographic history of the population as well as health reasons, such as population specific diseases. Some studies have sought to determine the relative contributions of the Turkic peoples of Central Asia, from where the Seljuk Turks began migrating to Anatolia after the Battle of Manzikert in 1071, which led to the establishment of the Anatolian Seljuk Sultanate in the late 11th century, and prior populations in the area who were culturally assimilated during the Seljuk and the Ottoman periods.
Genetic studies on Serbs show close affinity to other neighboring South Slavs.
The genetic history of Egypt reflects its geographical location at the crossroads of several major biocultural areas: North Africa, the Sahara, the Middle East, the Mediterranean and sub-Saharan Africa.
The Bulgarians are part of the Slavic ethnolinguistic group as a result of migrations of Slavic tribes to the region since the 6th century AD and the subsequent linguistic assimilation of other populations.
Population genetics is a scientific discipline which contributes to the examination of the human evolutionary and historical migrations. Particularly useful information is provided by the research of two uniparental markers within our genome, the Y-chromosome (Y-DNA) and mitochondrial DNA (mtDNA), as well as autosomal DNA. The data from Y-DNA and autosomal DNA suggests that the Croats mostly are descendants of the Slavs of the medieval migration period, according to mtDNA have genetic diversity which fits within a broader European maternal genetic landscape, and overall have a uniformity with other South Slavs from the territory of former Yugoslavia.
Various genetic studies on Filipinos have been performed, to analyze the population genetics of the various ethnic groups in the Philippines.
This article summarizes the genetic makeup and population history of East Asian peoples and their connection to genetically related populations, as well as Oceanians and partly, Central Asians and South Asians, which are collectively referred to as "East Eurasians" in population genomics.

The Scytho-Siberian world was an archaeological horizon that flourished across the entire Eurasian Steppe during the Iron Age, from approximately the 9th century BC to the 2nd century AD. It included the Scythian, Sauromatian and Sarmatian cultures of Eastern Europe, the Saka-Massagetae and Tasmola cultures of Central Asia, and the Aldy-Bel, Pazyryk and Tagar cultures of south Siberia.

In archaeogenetics, the term Western Steppe Herders (WSH), or Western Steppe Pastoralists, is the name given to a distinct ancestral component first identified in individuals from the Chalcolithic steppe around the turn of the 5th millennium BC, subsequently detected in several genetically similar or directly related ancient populations including the Khvalynsk, Sredny Stog, and Yamnaya cultures, and found in substantial levels in contemporary European, West Asian and South Asian populations. This ancestry is often referred to as Yamnaya ancestry, Yamnaya-related ancestry, Steppe ancestry or Steppe-related ancestry.

The genetic history of Africa summarizes the genetic makeup and population history of African populations in Africa, composed of the overall genetic history, including the regional genetic histories of North Africa, West Africa, East Africa, Central Africa, and Southern Africa, as well as the recent origin of modern humans in Africa. The Sahara served as a trans-regional passageway and place of dwelling for people in Africa during various humid phases and periods throughout the history of Africa.
Therefore, Siberian admixtures into Northeastern Europe likely began prior to 6.6 kya, coinciding with the expansion of Y-Chromosome haplogroup N1c1 among Siberians and Northeastern Europeans (7.1–4.9 kya). Since haplogroup N likely originated in Asia (Shi et al. 2013) and currently achieves its highest frequency among Siberian populations, its presence among Eastern Europeans likely reflects ancient gene flows from Siberia into Northeastern Europe.
Northeast Asians such as Oroqen, Mongolian, Hezhen and Daur (nomads who historically lived alongside Russians and Caucasians) inherited significantly more alleles from EUR: Mongolian 10.9 ± 0.1%, Oroqen 9.6 ± 0.2%, Daur 8.0 ± 0.2% and Hezhen 6.8 ± 0.2%.