In artificial intelligence, genetic programming (GP) is a technique of evolving programs, starting from a population of unfit programs, fit for a particular task by applying operations analogous to natural genetic processes to the population of programs.

In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to generate high-quality solutions to optimization and search problems by relying on biologically inspired operators such as mutation, crossover and selection. Some examples of GA applications include optimizing decision trees for better performance, solving sudoku puzzles, hyperparameter optimization, causal inference, etc.
In computational intelligence (CI), an evolutionary algorithm (EA) is a subset of evolutionary computation, a generic population-based metaheuristic optimization algorithm. An EA uses mechanisms inspired by biological evolution, such as reproduction, mutation, recombination, and selection. Candidate solutions to the optimization problem play the role of individuals in a population, and the fitness function determines the quality of the solutions. Evolution of the population then takes place after the repeated application of the above operators.
In computer science, evolutionary computation is a family of algorithms for global optimization inspired by biological evolution, and the subfield of artificial intelligence and soft computing studying these algorithms. In technical terms, they are a family of population-based trial and error problem solvers with a metaheuristic or stochastic optimization character.
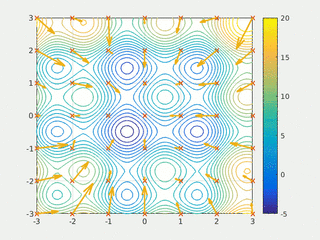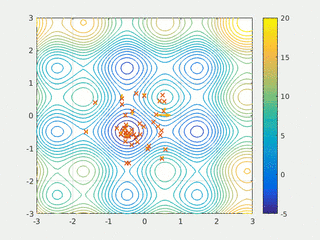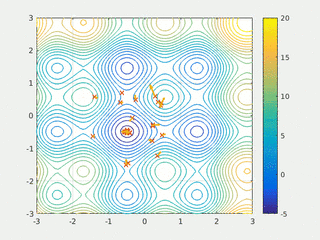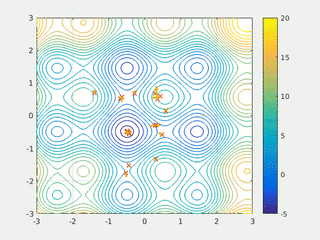
In computational science, particle swarm optimization (PSO) is a computational method that optimizes a problem by iteratively trying to improve a candidate solution with regard to a given measure of quality. It solves a problem by having a population of candidate solutions, here dubbed particles, and moving these particles around in the search-space according to simple mathematical formula over the particle's position and velocity. Each particle's movement is influenced by its local best known position, but is also guided toward the best known positions in the search-space, which are updated as better positions are found by other particles. This is expected to move the swarm toward the best solutions.
Neuroevolution, or neuro-evolution, is a form of artificial intelligence that uses evolutionary algorithms to generate artificial neural networks (ANN), parameters, and rules. It is most commonly applied in artificial life, general game playing and evolutionary robotics. The main benefit is that neuroevolution can be applied more widely than supervised learning algorithms, which require a syllabus of correct input-output pairs. In contrast, neuroevolution requires only a measure of a network's performance at a task. For example, the outcome of a game can be easily measured without providing labeled examples of desired strategies. Neuroevolution is commonly used as part of the reinforcement learning paradigm, and it can be contrasted with conventional deep learning techniques that use backpropagation with a fixed topology.
In genetic algorithms (GA), or more general, evolutionary algorithms (EA), a chromosome is a set of parameters which define a proposed solution of the problem that the evolutionary algorithm is trying to solve. The set of all solutions, also called individuals according to the biological model, is known as the population. The genome of an individual consists of one, more rarely of several, chromosomes and corresponds to the genetic representation of the task to be solved. A chromosome is composed of a set of genes, where a gene consists of one or more semantically connected parameters, which are often also called decision variables. They determine one or more phenotypic characteristics of the individual or at least have an influence on them. In the basic form of genetic algorithms, the chromosome is represented as a binary string, while in later variants and in EAs in general, a wide variety of other data structures are used.
In computer science and mathematical optimization, a metaheuristic is a higher-level procedure or heuristic designed to find, generate, tune, or select a heuristic that may provide a sufficiently good solution to an optimization problem or a machine learning problem, especially with incomplete or imperfect information or limited computation capacity. Metaheuristics sample a subset of solutions which is otherwise too large to be completely enumerated or otherwise explored. Metaheuristics may make relatively few assumptions about the optimization problem being solved and so may be usable for a variety of problems.
In computer programming, gene expression programming (GEP) is an evolutionary algorithm that creates computer programs or models. These computer programs are complex tree structures that learn and adapt by changing their sizes, shapes, and composition, much like a living organism. And like living organisms, the computer programs of GEP are also encoded in simple linear chromosomes of fixed length. Thus, GEP is a genotype–phenotype system, benefiting from a simple genome to keep and transmit the genetic information and a complex phenotype to explore the environment and adapt to it.
Linear genetic programming (LGP) is a particular method of genetic programming wherein computer programs in a population are represented as a sequence of instructions from an imperative programming language or machine language. The adjective "linear" stems from the fact that the sequence of instructions is normally executed in a linear fashion. Like in other programs, the data flow in LGP can be modeled as a graph that will visualize the potential multiple usage of register contents and the existence of structurally noneffective code (introns) which are two main differences of this genetic representation from the more common tree-based genetic programming (TGP) variant.
In natural evolution and artificial evolution the fitness of a schema is rescaled to give its effective fitness which takes into account crossover and mutation.
In computer science and operations research, Genetic fuzzy systems are fuzzy systems constructed by using genetic algorithms or genetic programming, which mimic the process of natural evolution, to identify its structure and parameter.

In computer science, the syntax of a computer language is the rules that define the combinations of symbols that are considered to be correctly structured statements or expressions in that language. This applies both to programming languages, where the document represents source code, and to markup languages, where the document represents data.
In computer programming, genetic representation is a way of presenting solutions/individuals in evolutionary computation methods. The term encompasses both the concrete data structures and data types used to realize the genetic material of the candidate solutions in the form of a genome, and the relationships between search space and problem space. In the simplest case, the search space corresponds to the problem space. The choice of problem representation is tied to the choice of genetic operators, both of which have a decisive effect on the efficiency of the optimization. Genetic representation can encode appearance, behavior, physical qualities of individuals. Difference in genetic representations is one of the major criteria drawing a line between known classes of evolutionary computation.
ECJ is a freeware evolutionary computation research system written in Java. It is a framework that supports a variety of evolutionary computation techniques, such as genetic algorithms, genetic programming, evolution strategies, coevolution, particle swarm optimization, and differential evolution. The framework models iterative evolutionary processes using a series of pipelines arranged to connect one or more subpopulations of individuals with selection, breeding (such as crossover, and mutation operators that produce new individuals. The framework is open source and is distributed under the Academic Free License. ECJ was created by Sean Luke, a computer science professor at George Mason University, and is maintained by Sean Luke and a variety of contributors.
GeneNetwork is a combined database and open-source bioinformatics data analysis software resource for systems genetics. This resource is used to study gene regulatory networks that link DNA sequence differences to corresponding differences in gene and protein expression and to variation in traits such as health and disease risk. Data sets in GeneNetwork are typically made up of large collections of genotypes and phenotypes from groups of individuals, including humans, strains of mice and rats, and organisms as diverse as Drosophila melanogaster, Arabidopsis thaliana, and barley. The inclusion of genotypes makes it practical to carry out web-based gene mapping to discover those regions of genomes that contribute to differences among individuals in mRNA, protein, and metabolite levels, as well as differences in cell function, anatomy, physiology, and behavior.
The MOEA Framework is an open-source evolutionary computation library for Java that specializes in multi-objective optimization. It supports a variety of multiobjective evolutionary algorithms (MOEAs), including genetic algorithms, genetic programming, grammatical evolution, differential evolution, and particle swarm optimization. As a result, it has been used to conduct numerous comparative studies to assess the efficiency, reliability, and controllability of state-of-the-art MOEAs.
Multi Expression Programming (MEP) is an evolutionary algorithm for generating mathematical functions describing a given set of data. MEP is a Genetic Programming variant encoding multiple solutions in the same chromosome. MEP representation is not specific. In the simplest variant, MEP chromosomes are linear strings of instructions. This representation was inspired by Three-address code. MEP strength consists in the ability to encode multiple solutions, of a problem, in the same chromosome. In this way, one can explore larger zones of the search space. For most of the problems this advantage comes with no running-time penalty compared with genetic programming variants encoding a single solution in a chromosome.
The following outline is provided as an overview of and topical guide to machine learning: