Related Research Articles
A Bayesian network is a probabilistic graphical model that represents a set of variables and their conditional dependencies via a directed acyclic graph (DAG). While it is one of several forms of causal notation, causal networks are special cases of Bayesian networks. Bayesian networks are ideal for taking an event that occurred and predicting the likelihood that any one of several possible known causes was the contributing factor. For example, a Bayesian network could represent the probabilistic relationships between diseases and symptoms. Given symptoms, the network can be used to compute the probabilities of the presence of various diseases.
A graphical model or probabilistic graphical model (PGM) or structured probabilistic model is a probabilistic model for which a graph expresses the conditional dependence structure between random variables. They are commonly used in probability theory, statistics—particularly Bayesian statistics—and machine learning.
The Bioinformatics Centre is an interdisciplinary research in bioinformatics at the University of Copenhagen (UCPH). The centre is housed in the Section for Computational and RNA Biology at the Department of Biology within the Faculty of Science.
Belief propagation, also known as sum–product message passing, is a message-passing algorithm for performing inference on graphical models, such as Bayesian networks and Markov random fields. It calculates the marginal distribution for each unobserved node, conditional on any observed nodes. Belief propagation is commonly used in artificial intelligence and information theory, and has demonstrated empirical success in numerous applications, including low-density parity-check codes, turbo codes, free energy approximation, and satisfiability.
An influence diagram (ID) is a compact graphical and mathematical representation of a decision situation. It is a generalization of a Bayesian network, in which not only probabilistic inference problems but also decision making problems can be modeled and solved.
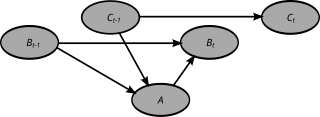
A dynamic Bayesian network (DBN) is a Bayesian network (BN) which relates variables to each other over adjacent time steps.

Daphne Koller is an Israeli-American computer scientist. She was a professor in the department of computer science at Stanford University and a MacArthur Foundation fellowship recipient. She is one of the founders of Coursera, an online education platform. Her general research area is artificial intelligence and its applications in the biomedical sciences. Koller was featured in a 2004 article by MIT Technology Review titled "10 Emerging Technologies That Will Change Your World" concerning the topic of Bayesian machine learning.
Variable-order Bayesian network (VOBN) models provide an important extension of both the Bayesian network models and the variable-order Markov models. VOBN models are used in machine learning in general and have shown great potential in bioinformatics applications. These models extend the widely used position weight matrix (PWM) models, Markov models, and Bayesian network (BN) models.

Krueppel-like factor 6 is a protein that in humans is encoded by the KLF6 gene.
Statistical relational learning (SRL) is a subdiscipline of artificial intelligence and machine learning that is concerned with domain models that exhibit both uncertainty and complex, relational structure. Typically, the knowledge representation formalisms developed in SRL use first-order logic to describe relational properties of a domain in a general manner and draw upon probabilistic graphical models to model the uncertainty; some also build upon the methods of inductive logic programming. Significant contributions to the field have been made since the late 1990s.
The Immunological Genome Project (ImmGen) is a collaborative scientific research project that is currently building a gene-expression database for all characterized immune cells in the mouse. The overarching goal of the project is to computationally reconstruct the gene regulatory network in immune cells
Probabilistic programming (PP) is a programming paradigm in which probabilistic models are specified and inference for these models is performed automatically. It represents an attempt to unify probabilistic modeling and traditional general purpose programming in order to make the former easier and more widely applicable. It can be used to create systems that help make decisions in the face of uncertainty.
Eran Segal is a computational biologist professor at the Weizmann Institute of Science. He works on developing quantitative models for all levels of gene regulation, including transcription, chromatin, and translation. Segal also works as an epidemiologist.

Aviv Regev is a computational biologist and systems biologist and Executive Vice President and Head of Genentech Research and Early Development in Genentech/Roche. She is a core member at the Broad Institute of MIT and Harvard and professor at the Department of Biology of the Massachusetts Institute of Technology. Regev is a pioneer of single cell genomics and of computational and systems biology of gene regulatory circuits. She founded and leads the Human Cell Atlas project, together with Sarah Teichmann.
Variable elimination (VE) is a simple and general exact inference algorithm in probabilistic graphical models, such as Bayesian networks and Markov random fields. It can be used for inference of maximum a posteriori (MAP) state or estimation of conditional or marginal distributions over a subset of variables. The algorithm has exponential time complexity, but could be efficient in practice for low-treewidth graphs, if the proper elimination order is used.

Dana Pe'er, Chair and Professor in Computational and Systems Biology Program at Sloan Kettering Institute is a researcher in computational systems biology. A Howard Hughes Medical Institute (HHMI) Investigator since 2021, she was previously a professor at Columbia Department of Biological Sciences. Pe'er's research focuses on understanding the organization, function and evolution of molecular networks, particularly how genetic variations alter the regulatory network and how these genetic variations can cause cancer.

Fred Marshall Winston is the John Emory Andrus Professor of Genetics in the Harvard Medical School Genetics Department, where he has been a member of the faculty since 1983. Research in his laboratory has focused on mechanisms of transcription and the regulation of chromatin structure in the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe. Dr. Winston served as the President of the Genetics Society of America in 2009 and has been elected to both the American Academy of Arts and Sciences (2009) and the National Academy of Sciences (2013).
Radford M. Neal is a professor emeritus at the Department of Statistics and Department of Computer Science at the University of Toronto, where he holds a research chair in statistics and machine learning.

Hanah Margalit is a Professor in the faculty of medicine at the Hebrew University of Jerusalem. Her research combines bioinformatics, computational biology and systems biology, specifically in the fields of gene regulation in bacteria and eukaryotes.

Michal Linial is a Professor of Biochemistry and Bioinformatics at the Hebrew University of Jerusalem (HUJI). Linial is the Director of The Sudarsky Center for Computational Biology at HUJI. Since 2015, she is head of the ELIXIR-Israel node.
References
- ↑ "Nir Friedman - Yad Hanadiv". Yadhanadiv.org.il. Archived from the original on 25 June 2016. Retrieved 25 May 2016.
- ↑ "Young HU biologist wins Australian award". The Jerusalem Post - JPost.com. Retrieved 25 May 2016.
- ↑ "Nir Friedman". Cs.huji.ac.il. Retrieved 25 May 2016.
- ↑ "Nir Friedman's Lab". Systemsbio.cs.huji.ac.il. Retrieved 25 May 2016.
- ↑ "Curriculum Vitae—Nir Friedman" (PDF). Cs.huji.ac.il. Retrieved 25 May 2016.
- ↑ "Archived copy". Archived from the original on April 25, 2012. Retrieved October 22, 2011.
{{cite web}}: CS1 maint: archived copy as title (link) - ↑ "The Rachel and Selim Benin School of Computer Science and Engineering". Cs.huji.ac.il. Retrieved 25 May 2016.
- ↑ "nir friedman - Google Scholar". Scholar.google.com. Retrieved 25 May 2016.
- ↑ Search Results for author Friedman N on PubMed .
- ↑ "Bayesian network classifiers". Cs.huji.ac.il. Retrieved 25 May 2016.
- ↑ "The Bayesian structural EM algorithm". Cs.huji.ac.il. Retrieved 25 May 2016.
- ↑ Friedman, N.; Linial, M.; Nachman, I.; Pe'er, D. (2000). "Using Bayesian Networks to Analyze Expression Data". Journal of Computational Biology. 7 (3–4): 601–620. CiteSeerX 10.1.1.191.139 . doi:10.1089/106652700750050961. PMID 11108481.
- ↑ Segal, E.; Shapira, M.; Regev, A.; Pe'er, D.; Botstein, D.; Koller, D.; Friedman, N. (2003). "Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data". Nature Genetics. 34 (2): 166–176. doi:10.1038/ng1165. PMID 12740579. S2CID 6146032.
- ↑ Friedman, N. (2004). "Inferring Cellular Networks Using Probabilistic Graphical Models". Science. 303 (5659): 799–805. Bibcode:2004Sci...303..799F. doi:10.1126/science.1094068. PMID 14764868. S2CID 6819971.
- ↑ Segal, E.; Friedman, N.; Koller, D.; Regev, A. (2004). "A module map showing conditional activity of expression modules in cancer". Nature Genetics. 36 (10): 1090–1098. doi: 10.1038/ng1434 . PMC 2271138 . PMID 15448693.
- ↑ "Nir Friedman". Cs.huji.ac.il. Retrieved 25 May 2016.
- ↑ Daphne Koller and Nir Friedman (2009). Probabilistic Graphical Models. MIT Press. ISBN 978-0-262-01319-2.
- ↑ "The Alexander Silberman Institute of Life Sciences". Bio.huji.ac.il. Archived from the original on 2016-04-26. Retrieved 2016-05-25.
- ↑ "Robotic Facility". Systemsbio.cs.huji.ac.il. Retrieved 25 May 2016.