
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then gene silence said mRNA molecules by one or more of the following processes:
- Cleavage of mRNA strand into two pieces,
- Destabilization of mRNA by shortening its poly(A) tail, or
- Translation of mRNA into proteins.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, and ultimately affect a phenotype. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication.

A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.

Functional genomics is a field of molecular biology that attempts to describe gene functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects. Functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "candidate-gene" approach.
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).
RNA silencing or RNA interference refers to a family of gene silencing effects by which gene expression is negatively regulated by non-coding RNAs such as microRNAs. RNA silencing may also be defined as sequence-specific regulation of gene expression triggered by double-stranded RNA (dsRNA). RNA silencing mechanisms are conserved among most eukaryotes. The most common and well-studied example is RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA).
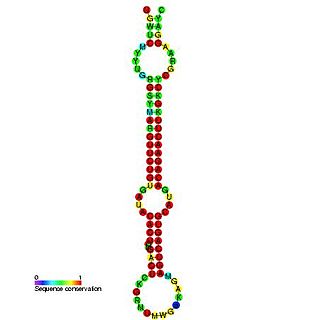
In molecular biology, miR-148 is a microRNA whose expression has been demonstrated in human, mouse, rat and zebrafish. miR-148 has also been predicted in chicken.
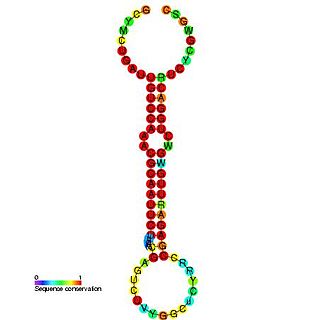
In molecular biology, the microRNA miR-219 was predicted in vertebrates by conservation between human, mouse and pufferfish and cloned in pufferfish. It was later predicted and confirmed experimentally in Drosophila. Homologs of miR-219 have since been predicted or experimentally confirmed in a wide range of species, including the platyhelminth Schmidtea mediterranea, several arthropod species and a wide range of vertebrates. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.
Anders Krogh is a bioinformatician at the University of Copenhagen, where he leads the university's bioinformatics center. He is known for his pioneering work on the use of hidden Markov models in bioinformatics, and is co-author of a widely used textbook in bioinformatics. In addition, he also co-authored one of the early textbooks on neural networks. His current research interests include promoter analysis, non-coding RNA, gene prediction and protein structure prediction.
Post-transcriptional regulation is the control of gene expression at the RNA level. It occurs once the RNA polymerase has been attached to the gene's promoter and is synthesizing the nucleotide sequence. Therefore, as the name indicates, it occurs between the transcription phase and the translation phase of gene expression. These controls are critical for the regulation of many genes across human tissues. It also plays a big role in cell physiology, being implicated in pathologies such as cancer and neurodegenerative diseases.
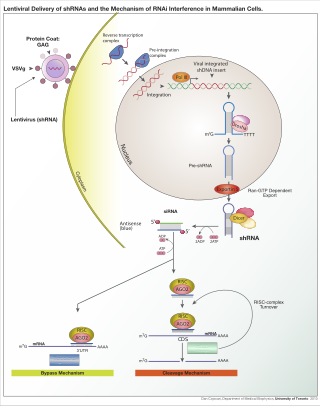
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.

Steven Elliot Brenner is a professor at the Department of Plant and Microbial Biology at the University of California Berkeley, adjunct professor at the Department of Bioengineering and Therapeutic Sciences at the University of California, and San Francisco Faculty scientist, Physical Biosciences at the Lawrence Berkeley National Laboratory.

Chris Sander is a computational biologist based at the Dana-Farber Cancer Center and Harvard Medical School. Previously he was chair of the Computational Biology Programme at the Memorial Sloan–Kettering Cancer Center in New York City. In 2015, he moved his lab to the Dana–Farber Cancer Institute and the Cell Biology Department at Harvard Medical School.

Burkhard Rost is a scientist leading the Department for Computational Biology & Bioinformatics at the Faculty of Informatics of the Technical University of Munich (TUM). Rost chairs the Study Section Bioinformatics Munich involving the TUM and the Ludwig Maximilian University of Munich (LMU) in Munich. From 2007-2014 Rost was President of the International Society for Computational Biology (ISCB).

Gary Stormo is an American geneticist and currently Joseph Erlanger Professor in the Department of Genetics and the Center for Genome Sciences and Systems Biology at Washington University School of Medicine in St Louis. He is considered one of the pioneers of bioinformatics and genomics. His research combines experimental and computational approaches in order to identify and predict regulatory sequences in DNA and RNA, and their contributions to the regulatory networks that control gene expression.

Alexander George Bateman is a computational biologist and Head of Protein Sequence Resources at the European Bioinformatics Institute (EBI), part of the European Molecular Biology Laboratory (EMBL) in Cambridge, UK. He has led the development of the Pfam biological database and introduced the Rfam database of RNA families. He has also been involved in the use of Wikipedia for community-based annotation of biological databases.
Direct coupling analysis or DCA is an umbrella term comprising several methods for analyzing sequence data in computational biology. The common idea of these methods is to use statistical modeling to quantify the strength of the direct relationship between two positions of a biological sequence, excluding effects from other positions. This contrasts usual measures of correlation, which can be large even if there is no direct relationship between the positions. Such a direct relationship can for example be the evolutionary pressure for two positions to maintain mutual compatibility in the biomolecular structure of the sequence, leading to molecular coevolution between the two positions.

Hanah Margalit is a Professor in the faculty of medicine at the Hebrew University of Jerusalem. Her research combines bioinformatics, computational biology and systems biology, specifically in the fields of gene regulation in bacteria and eukaryotes.
Rita Casadio is an Adjunct Professor of Biochemistry/Biophysics in the Department of Pharmacy and Biotechnology at the University of Bologna.














