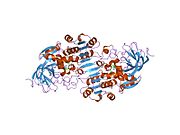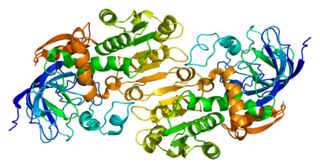
Alcohol dehydrogenases (ADH) (EC 1.1.1.1) are a group of dehydrogenase enzymes that occur in many organisms and facilitate the interconversion between alcohols and aldehydes or ketones with the reduction of nicotinamide adenine dinucleotide (NAD+) to NADH. In humans and many other animals, they serve to break down alcohols that are otherwise toxic, and they also participate in the generation of useful aldehyde, ketone, or alcohol groups during the biosynthesis of various metabolites. In yeast, plants, and many bacteria, some alcohol dehydrogenases catalyze the opposite reaction as part of fermentation to ensure a constant supply of NAD+.

Acetaldehyde dehydrogenases are dehydrogenase enzymes which catalyze the conversion of acetaldehyde into acetyl-CoA. This can be summarized as follows:

Alcohol tolerance refers to the bodily responses to the functional effects of ethanol in alcoholic beverages. This includes direct tolerance, speed of recovery from insobriety and resistance to the development of alcohol use disorder.
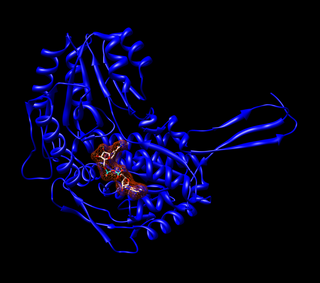
Aldehyde dehydrogenases are a group of enzymes that catalyse the oxidation of aldehydes. They convert aldehydes to carboxylic acids. The oxygen comes from a water molecule. To date, nineteen ALDH genes have been identified within the human genome. These genes participate in a wide variety of biological processes including the detoxification of exogenously and endogenously generated aldehydes.
Ethanol, an alcohol found in nature and in alcoholic drinks, is metabolized through a complex catabolic metabolic pathway. In humans, several enzymes are involved in processing ethanol first into acetaldehyde and further into acetic acid and acetyl-CoA. Once acetyl-CoA is formed, it becomes a substrate for the citric acid cycle ultimately producing cellular energy and releasing water and carbon dioxide. Due to differences in enzyme presence and availability, human adults and fetuses process ethanol through different pathways. Gene variation in these enzymes can lead to variation in catalytic efficiency between individuals. The liver is the major organ that metabolizes ethanol due to its high concentration of these enzymes.

Aldehyde dehydrogenase, mitochondrial is an enzyme that in humans is encoded by the ALDH2 gene located on chromosome 12. ALDH2 belongs to the aldehyde dehydrogenase family of enzymes. Aldehyde dehydrogenase is the second enzyme of the major oxidative pathway of alcohol metabolism. ALDH2 has a low Km for acetaldehyde, and is localized in mitochondrial matrix. The other liver isozyme, ALDH1, localizes to the cytosol.
In enzymology, a retinol dehydrogenase (RDH) (EC 1.1.1.105) is an enzyme that catalyzes the chemical reaction
In enzymology, a S-(hydroxymethyl)glutathione dehydrogenase (EC 1.1.1.284) is an enzyme that catalyzes the chemical reaction
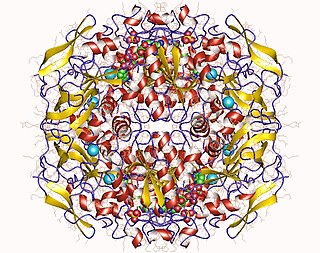
In enzymology, a formaldehyde dehydrogenase (EC 1.2.1.46) is an enzyme that catalyzes the chemical reaction

Alcohol dehydrogenase 1C is an enzyme that in humans is encoded by the ADH1C gene.

In enzymology, a formate—tetrahydrofolate ligase is an enzyme that catalyzes the chemical reaction

Alcohol dehydrogenase class-3 is an enzyme that in humans is encoded by the ADH5 gene.

Alcohol dehydrogenase 4 is an enzyme that in humans is encoded by the ADH4 gene.

Alcohol dehydrogenase 1A is an enzyme that in humans is encoded by the ADH1A gene.

Alcohol dehydrogenase class 4 mu/sigma chain is an enzyme that in humans is encoded by the ADH7 gene.

Gamma-aminobutyric acid receptor subunit alpha-2 is a protein in humans that is encoded by the GABRA2 gene.

Alcohol dehydrogenase 6 is an enzyme that in humans is encoded by the ADH6 gene.

GABAA receptor-γ3, also known as GABRG3, is a protein which in humans is encoded by the GABRG3 gene.

The short-term effects of alcohol consumption range from a decrease in anxiety and motor skills and euphoria at lower doses to intoxication (drunkenness), to stupor, unconsciousness, anterograde amnesia, and central nervous system depression at higher doses. Cell membranes are highly permeable to alcohol, so once it is in the bloodstream, it can diffuse into nearly every cell in the body.

Alcohol intolerance is due to a genetic polymorphism of the aldehyde dehydrogenase enzyme, which is responsible for the metabolism of acetaldehyde. This polymorphism is most often reported in patients of East Asian descent. Alcohol intolerance may also be an associated side effect of certain drugs such as disulfiram, metronidazole, or nilutamide. Skin flushing and nasal congestion are the most common symptoms of intolerance after alcohol ingestion. It may also be characterized as intolerance causing hangover symptoms similar to the "disulfiram-like reaction" of aldehyde dehydrogenase deficiency or chronic fatigue syndrome. Severe pain after drinking alcohol may indicate a more serious underlying condition.