Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus. It is a parasexual mode of reproduction in bacteria.
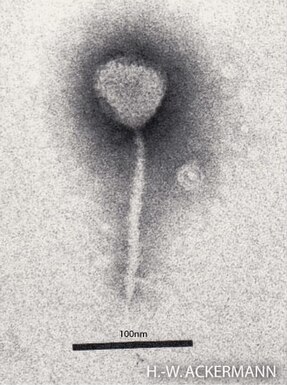
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

Escherichia coli, also known as E. coli, is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes (EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. The harmless strains are part of the normal microbiota of the gut, and can benefit their hosts by producing vitamin K2, and preventing colonisation of the intestine with pathogenic bacteria, having a mutualistic relationship. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

Shiga toxins are a family of related toxins with two major groups, Stx1 and Stx2, expressed by genes considered to be part of the genome of lambdoid prophages. The toxins are named after Kiyoshi Shiga, who first described the bacterial origin of dysentery caused by Shigella dysenteriae. Shiga-like toxin (SLT) is a historical term for similar or identical toxins produced by Escherichia coli. The most common sources for Shiga toxin are the bacteria S. dysenteriae and some serotypes of Escherichia coli (STEC), which includes serotypes O157:H7, and O104:H4.
DNA topoisomerases are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in DNA arise due to the intertwined nature of its double-helical structure, which, for example, can lead to overwinding of the DNA duplex during DNA replication and transcription. If left unchanged, this torsion would eventually stop the DNA or RNA polymerases involved in these processes from continuing along the DNA helix. A second topological challenge results from the linking or tangling of DNA during replication. Left unresolved, links between replicated DNA will impede cell division. The DNA topoisomerases prevent and correct these types of topological problems. They do this by binding to DNA and cutting the sugar-phosphate backbone of either one or both of the DNA strands. This transient break allows the DNA to be untangled or unwound, and, at the end of these processes, the DNA backbone is resealed. Since the overall chemical composition and connectivity of the DNA do not change, the DNA substrate and product are chemical isomers, differing only in their topology.
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

FtsZ is a protein encoded by the ftsZ gene that assembles into a ring at the future site of bacterial cell division. FtsZ is a prokaryotic homologue of the eukaryotic protein tubulin. The initials FtsZ mean "Filamenting temperature-sensitive mutant Z." The hypothesis was that cell division mutants of E. coli would grow as filaments due to the inability of the daughter cells to separate from one another. FtsZ is found in almost all bacteria, many archaea, all chloroplasts and some mitochondria, where it is essential for cell division. FtsZ assembles the cytoskeletal scaffold of the Z ring that, along with additional proteins, constricts to divide the cell in two.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages. This can eventually lead to malignant tumors, or cancer as per the two hit hypothesis.

Bleomycin is a medication used to treat cancer. This includes Hodgkin's lymphoma, non-Hodgkin's lymphoma, testicular cancer, ovarian cancer, and cervical cancer among others. Typically used with other cancer medications, it can be given intravenously, by injection into a muscle or under the skin. It may also be administered inside the chest to help prevent the recurrence of a fluid around the lung due to cancer; however talc is better for this.

RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.

Lysogeny, or the lysogenic cycle, is one of two cycles of viral reproduction. Lysogeny is characterized by integration of the bacteriophage nucleic acid into the host bacterium's genome or formation of a circular replicon in the bacterial cytoplasm. In this condition the bacterium continues to live and reproduce normally, while the bacteriophage lies in a dormant state in the host cell. The genetic material of the bacteriophage, called a prophage, can be transmitted to daughter cells at each subsequent cell division, and later events can release it, causing proliferation of new phages via the lytic cycle. Lysogenic cycles can also occur in eukaryotes, although the method of DNA incorporation is not fully understood. For instance the AIDS viruses can either infect humans lytically, or lay dormant (lysogenic) as part of the infected cells' genome, keeping the ability to return to lysis at a later time. The rest of this article is about lysogeny in bacterial hosts.
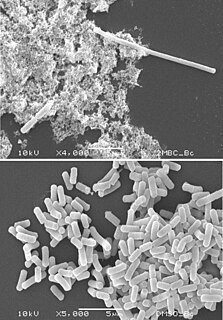
Filamentation, also termed conditional filamentation, is the anomalous growth of certain bacteria, such as Escherichia coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies. In the absence of antibiotics or other stressors, filamentation occurs at a low frequency in bacterial populations. The increased cell length can protecting bacteria from protozoan predation and neutrophil phagocytosis by making ingestion of cells more difficult. Filamentation is also thought to protect bacteria from antibiotics, and is associated with other aspects of bacterial virulence such as biofilm formation. The number and length of filaments within a bacterial population increases when the bacteria are treated with various chemical and physical agents. Some of the key genes involved in filamentation in E. coli include sulA and minCD.
Recombineering is a genetic and molecular biology technique based on homologous recombination systems, as opposed to the older/more common method of using restriction enzymes and ligases to combine DNA sequences in a specified order. Recombineering is widely used for bacterial genetics, in the generation of target vectors for making a conditional mouse knockout, and for modifying DNA of any source often contained on a bacterial artificial chromosome (BAC), among other applications.
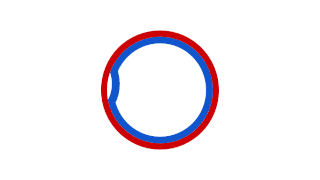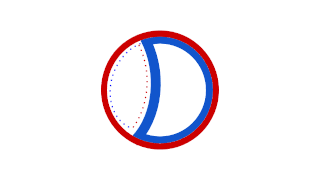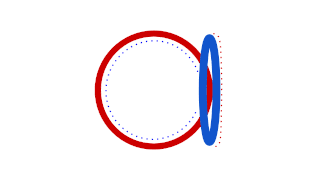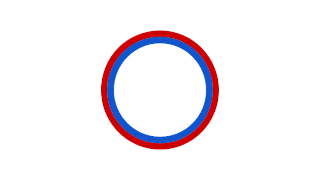
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.
A P1-derived artificial chromosome, or PAC, is a DNA construct derived from the DNA of P1 bacteriophages and Bacterial artificial chromosome. It can carry large amounts of other sequences for a variety of bioengineering purposes in bacteria. It is one type of the efficient cloning vector used to clone DNA fragments in Escherichia coli cells.
Persister cells are subpopulations of cells that resist treatment, and become antimicrobial tolerant by changing to a state of dormancy or quiescence. Persister cells in their dormancy do not divide. The tolerance shown in persister cells differs from antimicrobial resistance in that the tolerance is not inherited and is reversible. When treatment has stopped the state of dormancy can be reversed and the cells can reactivate and multiply. Most persister cells are bacterial, and there are also fungal persister cells, yeast persister cells, and cancer persister cells that show tolerance for cancer drugs.
Zygotic induction occurs when a bacterial cell carrying the silenced DNA of a bacterial virus in its chromosome transfers the viral DNA along with its own DNA to another bacterial cell lacking the virus, causing the recipient of the DNA to break open. In the donor cell, a repressor protein encoded by the prophage keeps the viral genes turned off so that virus is not produced. When DNA is transferred to the recipient cell by conjugation, the viral genes in the transferred DNA are immediately turned on because the recipient cell lacks the repressor. As a result, many virus are made in the recipient cell, and lysis eventually occurs to release the new virus.

Escherichia coli is a gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but pathogenic varieties cause serious food poisoning, septic shock, meningitis, or urinary tract infections in humans. Unlike normal flora E. coli, the pathogenic varieties produce toxins and other virulence factors that enable them to reside in parts of the body normally not inhabited by E. coli, and to damage host cells. These pathogenic traits are encoded by virulence genes carried only by the pathogens.

In molecular biology, the Fic/DOC protein family is a family of proteins which catalyzes the post-translational modification of proteins using phosphate-containing compound as a substrate. Fic domain proteins typically use ATP as a co-factor, but in some cases GTP or UTP is used. Post-translational modification performed by Fic domains is usually NMPylation, however they also catalyze phosphorylation and phosphocholine transfer. This family contains a central conserved motif HPFX[D/E]GNGR in most members and it carries the invariant catalytic histidine. Fic domain was found in bacteria, eukaryotes and archaea and can be found organized in almost hundred different multi-domain assemblies.
The CTXφ bacteriophage is a filamentous bacteriophage. It is a positive-strand DNA virus with single-stranded DNA (ssDNA).












