Cytochromes are redox-active proteins containing a heme, with a central iron (Fe) atom at its core, as a cofactor. They are involved in electron transport chain and redox catalysis. They are classified according to the type of heme and its mode of binding. Four varieties are recognized by the International Union of Biochemistry and Molecular Biology (IUBMB), cytochromes a, cytochromes b, cytochromes c and cytochrome d.

The enzyme cytochrome c oxidase or Complex IV, is a large transmembrane protein complex found in bacteria, archaea, and mitochondria of eukaryotes.
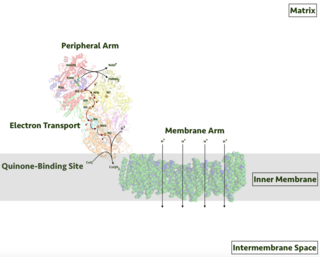
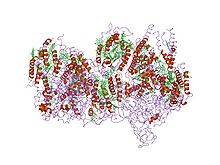
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.

Plastoquinone (PQ) is an isoprenoid quinone molecule involved in the electron transport chain in the light-dependent reactions of photosynthesis. The most common form of plastoquinone, known as PQ-A or PQ-9, is a 2,3-dimethyl-1,4-benzoquinone molecule with a side chain of nine isoprenyl units. There are other forms of plastoquinone, such as ones with shorter side chains like PQ-3 as well as analogs such as PQ-B, PQ-C, and PQ-D, which differ in their side chains. The benzoquinone and isoprenyl units are both nonpolar, anchoring the molecule within the inner section of a lipid bilayer, where the hydrophobic tails are usually found.

Photosystems are functional and structural units of protein complexes involved in photosynthesis. Together they carry out the primary photochemistry of photosynthesis: the absorption of light and the transfer of energy and electrons. Photosystems are found in the thylakoid membranes of plants, algae, and cyanobacteria. These membranes are located inside the chloroplasts of plants and algae, and in the cytoplasmic membrane of photosynthetic bacteria. There are two kinds of photosystems: PSI and PSII.

Photosystem II is the first protein complex in the light-dependent reactions of oxygenic photosynthesis. It is located in the thylakoid membrane of plants, algae, and cyanobacteria. Within the photosystem, enzymes capture photons of light to energize electrons that are then transferred through a variety of coenzymes and cofactors to reduce plastoquinone to plastoquinol. The energized electrons are replaced by oxidizing water to form hydrogen ions and molecular oxygen.

Photosystem I is one of two photosystems in the photosynthetic light reactions of algae, plants, and cyanobacteria. Photosystem I is an integral membrane protein complex that uses light energy to catalyze the transfer of electrons across the thylakoid membrane from plastocyanin to ferredoxin. Ultimately, the electrons that are transferred by Photosystem I are used to produce the moderate-energy hydrogen carrier NADPH. The photon energy absorbed by Photosystem I also produces a proton-motive force that is used to generate ATP. PSI is composed of more than 110 cofactors, significantly more than Photosystem II.

The cytochrome b6f complex is an enzyme found in the thylakoid membrane in chloroplasts of plants, cyanobacteria, and green algae, that catalyzes the transfer of electrons from plastoquinol to plastocyanin. The reaction is analogous to the reaction catalyzed by cytochrome bc1 of the mitochondrial electron transport chain. During photosynthesis, the cytochrome b6f complex is one step along the chain that transfers electrons from Photosystem II to Photosystem I, and at the same time pumps protons into the thylakoid space, contributing to the generation of an electrochemical (energy) gradient that is later used to synthesize ATP from ADP.

An electrochemical gradient is a gradient of electrochemical potential, usually for an ion that can move across a membrane. The gradient consists of two parts:

Biliverdin reductase (BVR) is an enzyme found in all tissues under normal conditions, but especially in reticulo-macrophages of the liver and spleen. BVR facilitates the conversion of biliverdin to bilirubin via the reduction of a double-bond between the second and third pyrrole ring into a single-bond.

A photosynthetic reaction center is a complex of several proteins, pigments and other co-factors that together execute the primary energy conversion reactions of photosynthesis. Molecular excitations, either originating directly from sunlight or transferred as excitation energy via light-harvesting antenna systems, give rise to electron transfer reactions along the path of a series of protein-bound co-factors. These co-factors are light-absorbing molecules (also named chromophores or pigments) such as chlorophyll and pheophytin, as well as quinones. The energy of the photon is used to excite an electron of a pigment. The free energy created is then used, via a chain of nearby electron acceptors, for a transfer of hydrogen atoms (as protons and electrons) from H2O or hydrogen sulfide towards carbon dioxide, eventually producing glucose. These electron transfer steps ultimately result in the conversion of the energy of photons to chemical energy.
P700, or photosystem I primary donor, is the reaction-center chlorophyll a molecular dimer associated with photosystem I in plants, algae, and cyanobacteria.

Photoinhibition is light-induced reduction in the photosynthetic capacity of a plant, alga, or cyanobacterium. Photosystem II (PSII) is more sensitive to light than the rest of the photosynthetic machinery, and most researchers define the term as light-induced damage to PSII. In living organisms, photoinhibited PSII centres are continuously repaired via degradation and synthesis of the D1 protein of the photosynthetic reaction center of PSII. Photoinhibition is also used in a wider sense, as dynamic photoinhibition, to describe all reactions that decrease the efficiency of photosynthesis when plants are exposed to light.

Stigmatellin is a potent inhibitor of the quinol oxidation (Qo) site of the cytochrome bc1 complex in mitochondria and the cytochrome b6f complex of thylakoid membranes. At higher concentrations, stigmatellin also inhibits Complex I, as a "Class B" inhibitor of that enzyme.

Photosynthetic reaction centre proteins are main protein components of photosynthetic reaction centres (RCs) of bacteria and plants. They are transmembrane proteins embedded in the chloroplast thylakoid or bacterial cell membrane.

Cytochromes c cytochromes, or heme-containing proteins, that have heme C covalently attached to the peptide backbone via one or two thioether bonds. These bonds are in most cases part of a specific Cys-X-X-Cys-His (CXXCH) binding motif, where X denotes a miscellaneous amino acid. Two thioether bonds of cysteine residues bind to the vinyl sidechains of heme, and the histidine residue coordinates one axial binding site of the heme iron. Less common binding motifs can include a single thioether linkage, a lysine or a methionine instead of the axial histidine or a CXnCH binding motif with n>2. The second axial site of the iron can be coordinated by amino acids of the protein, substrate molecules or water. Cytochromes c possess a wide range of properties and function as electron transfer proteins or catalyse chemical reactions involving redox processes. A prominent member of this family is mitochondrial cytochrome c.

Light-dependent reactions is jargon for certain photochemical reactions that are involved in photosynthesis, the main process by which plants acquire energy. There are two light dependent reactions, the first occurs at photosystem II (PSII) and the second occurs at photosystem I (PSI),

Flavocytochrome c sulfide dehydrogenase, also known as Sulfide-cytochrome-c reductase (flavocytochrome c) (EC 1.8.2.3), is an enzyme with systematic name hydrogen-sulfide:flavocytochrome c oxidoreductase. It is found in sulfur-oxidising bacteria such as the purple phototrophic bacteria Allochromatium vinosum. This enzyme catalyses the following chemical reaction:
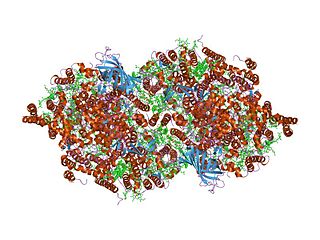
In molecular biology, the PsbZ (Ycf9) is a protein domain, which is low in molecular weight. It is a transmembrane protein and therefore is located in the thylakoid membrane of chloroplasts in cyanobacteria and plants. More specifically, it is located in Photosystem II (PSII) and in the light-harvesting complex II (LHCII). Ycf9 acts as a structural linker, that stabilises the PSII-LHCII supercomplexes. Moreover, the supercomplex fails to form in PsbZ-deficient mutants, providing further evidence to suggest Ycf9's role as a structural linker. This may be caused by a marked decrease in two LHCII antenna proteins, CP26 and CP29, found in PsbZ-deficient mutants, which result in structural changes, as well as functional modifications in PSII.

Cytochrome d, previously known as cytochrome a2, is a name for all cytochromes that contain heme D as a cofactor. Two unrelated classes of cytochrome d are known: Cytochrome bd, an enzyme that generates a charge across the membrane so that protons will move, and cytochrome cd1, a nitrite reductase.