Heme or haem is a substance precursive to hemoglobin, which is necessary to bind oxygen in the bloodstream. Haem is biosynthesized in both the bone marrow and the liver.

The globins are a superfamily of heme-containing globular proteins, involved in binding and/or transporting oxygen. These proteins all incorporate the globin fold, a series of eight alpha helical segments. Two prominent members include myoglobin and hemoglobin. Both of these proteins reversibly bind oxygen via a heme prosthetic group. They are widely distributed in many organisms.

Lignin is a class of complex organic polymers that form key structural materials in the support tissues of vascular plants and some algae. Lignins are particularly important in the formation of cell walls, especially in wood and bark, because they lend rigidity and do not rot easily. Chemically, lignins are cross-linked phenolic polymers.

Heme B or haem B is the most abundant heme. Hemoglobin and myoglobin are examples of oxygen transport proteins that contain heme B. The peroxidase family of enzymes also contain heme B. The COX-1 and COX-2 enzymes (cyclooxygenase) of recent fame, also contain heme B at one of two active sites.
Lignin-modifying enzymes (LMEs) are various types of enzymes produced by fungi and bacteria that catalyze the breakdown of lignin, a biopolymer commonly found in the cell walls of plants. The terms ligninases and lignases are older names for the same class, but the name "lignin-modifying enzymes" is now preferred, given that these enzymes are not hydrolytic but rather oxidative by their enzymatic mechanisms. LMEs include peroxidases, such as lignin peroxidase, manganese peroxidase, versatile peroxidase, and many phenoloxidases of the laccase type.
Prephenate dehydrogenase is an enzyme found in the shikimate pathway, and helps catalyze the reaction from prephenate to tyrosine.
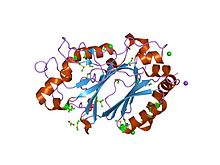
In enzymology, a manganese peroxidase (EC 1.11.1.13) is an enzyme that catalyzes the chemical reaction
In enzymology, a holocytochrome-c synthase is an enzyme that catalyzes the chemical reaction
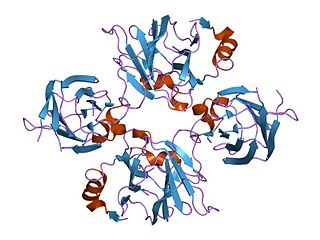
Allantoicase is an enzyme (EC 3.5.3.4) that in humans is encoded by the ALLC gene. Allantoicase catalyzes the chemical reaction

Cytochromes c cytochromes, or heme-containing proteins, that have heme C covalently attached to the peptide backbone via one or two thioether bonds. These bonds are in most cases part of a specific Cys-X-X-Cys-His (CXXCH) binding motif, where X denotes a miscellaneous amino acid. Two thioether bonds of cysteine residues bind to the vinyl sidechains of heme, and the histidine residue coordinates one axial binding site of the heme iron. Less common binding motifs can include a single thioether linkage, a lysine or a methionine instead of the axial histidine or a CXnCH binding motif with n>2. The second axial site of the iron can be coordinated by amino acids of the protein, substrate molecules or water. Cytochromes c possess a wide range of properties and function as electron transfer proteins or catalyse chemical reactions involving redox processes. A prominent member of this family is mitochondrial cytochrome c.

Dual oxidase 1, also known as DUOX1 or ThOX1, is an enzyme which in humans is encoded by the DUOX1 gene. DUOX1 was first identified in the mammalian thyroid gland. In humans, two isoforms are found; hDUOX1 and hDUOX2. Human DUOX protein localization is not exclusive to thyroid tissue; hDUOX1 is prominent in airway epithelial cells and hDUOX2 in the salivary glands and gastrointestinal tract.

Scavenger mRNA-decapping enzyme DcpS is a protein that in humans is encoded by the DCPS gene.
A domain of unknown function (DUF) is a protein domain that has no characterised function. These families have been collected together in the Pfam database using the prefix DUF followed by a number, with examples being DUF2992 and DUF1220. As of 2019, there are almost 4,000 DUF families within the Pfam database representing over 22% of known families. Some DUFs are not named using the nomenclature due to popular usage but are nevertheless DUFs.
Haem peroxidases (or heme peroxidases) are haem-containing enzymes that use hydrogen peroxide as the electron acceptor to catalyse a number of oxidative reactions. Most haem peroxidases follow the reaction scheme:
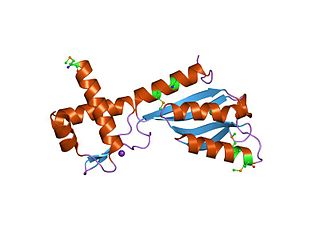
In molecular biology, the 3H domain is a protein domain named after its three highly conserved histidine residues. The 3H domain appears to be a smarr molecure-binding domain, based on its occurrence with other domains. Several proteins carrying this domain are transcriptional regulators from the biotin repressor family. The transcription regulator TM1602 from Thermotoga maritima is a DNA-binding protein thought to belong to a family of de novo NAD synthesis pathway regulators. TM1602 has an N-terminal DNA-binding domain and a C-terminal 3H regulatory domain. The N-terminal domain appears to bind to the NAD promoter region and repress the de novo NAD biosynthesis operon, while the C-terminal 3H domain may bind to nicotinamide, nicotinic acid, or other substrate/products. The 3H domain has a 2-layer alpha/beta sandwich fold.

In molecular biology, the di-haem cytochrome c peroxidase family is a group of distinct cytochrome c peroxidases (CCPs) that contain two haem groups. Similar to other cytochrome c peroxidases, they reduce hydrogen peroxide to water using c-type haem as an oxidizable substrate. However, since they possess two, instead of one, haem prosthetic groups, this family of bacterial CCPs reduce hydrogen peroxide without the need to generate semi-stable free radicals. The two haem groups have significantly different redox potentials. The high potential haem feeds electrons from electron shuttle proteins to the low potential haem, where peroxide is reduced. The CCP protein itself is structured into two domains, each containing one c-type haem group, with a calcium-binding site at the domain interface. This family also includes MauG proteins, whose similarity to di-haem CCP was previously recognised.

In molecular biology, enzymes containing the cyclodeaminase domain function in channeling one-carbon units to the folate pool. In most cases, this domain acts as a formimidoyltetrahydrofolate cyclodeaminase, which catalyses the cyclisation of formimidoyltetrahydrofolate to methenyltetrahydrofolate as shown in reaction (1). In the methylotrophic bacterium Methylobacterium extorquens, however, it acts as a methenyltetrahydrofolate cyclohydrolase, which catalyses the interconversion of formyltetrahydrofolate and methylenetetrahydrofolate, as shown in reaction (2).
Dye-decolorizing peroxidase (EC 1.11.1.19, DyP, DyP-type peroxidase) is an enzyme with systematic name Reactive-Blue-5:hydrogen-peroxide oxidoreductase. This enzyme catalyses the following chemical reaction

Eosinophil peroxidase is an enzyme found within the eosinophil granulocytes, innate immune cells of humans and mammals. This oxidoreductase protein is encoded by the gene EPX, expressed within these myeloid cells. EPO shares many similarities with its orthologous peroxidases, myeloperoxidase (MPO), lactoperoxidase (LPO), and thyroid peroxidase (TPO). The protein is concentrated in secretory granules within eosinophils. Eosinophil peroxidase is a heme peroxidase, its activities including the oxidation of halide ions to bacteriocidal reactive oxygen species, the cationic disruption of bacterial cell walls, and the post-translational modification of protein amino acid residues.

Histidine phosphotransfer domains and histidine phosphotransferases are protein domains involved in the "phosphorelay" form of two-component regulatory systems. These proteins possess a phosphorylatable histidine residue and are responsible for transferring a phosphoryl group from an aspartate residue on an intermediate "receiver" domain, typically part of a hybrid histidine kinase, to an aspartate on a final response regulator.