
Molybdenum is a chemical element; it has symbol Mo and atomic number 42. The name derived from Ancient Greek Μόλυβδος molybdos, meaning lead, since its ores were confused with lead ores. Molybdenum minerals have been known throughout history, but the element was discovered in 1778 by Carl Wilhelm Scheele. The metal was first isolated in 1781 by Peter Jacob Hjelm.
Nitrogen fixation is a chemical process by which molecular nitrogen (N
2), which has a strong triple covalent bond, is converted into ammonia (NH
3) or related nitrogenous compounds, typically in soil or aquatic systems but also in industry. The nitrogen in air is molecular dinitrogen, a relatively nonreactive molecule that is metabolically useless to all but a few microorganisms. Biological nitrogen fixation or diazotrophy is an important microbe-mediated process that converts dinitrogen (N2) gas to ammonia (NH3) using the nitrogenase protein complex (Nif).
Metalloprotein is a generic term for a protein that contains a metal ion cofactor. A large proportion of all proteins are part of this category. For instance, at least 1000 human proteins contain zinc-binding protein domains although there may be up to 3000 human zinc metalloproteins.
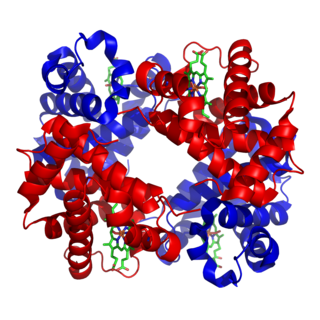
Nitrogenases are enzymes (EC 1.18.6.1EC 1.19.6.1) that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase.
Azotobacter vinelandii is Gram-negative diazotroph that can fix nitrogen while grown aerobically. These bacteria are easily cultured and grown.
Iron–sulfur proteins are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, forming dinitrosyl iron complexes. In most Fe–S proteins, the terminal ligands on Fe are thiolate, but exceptions exist.

Molybdopterins are a class of cofactors found in most molybdenum-containing and all tungsten-containing enzymes. Synonyms for molybdopterin are: MPT and pyranopterin-dithiolate. The nomenclature for this biomolecule can be confusing: Molybdopterin itself contains no molybdenum; rather, this is the name of the ligand that will bind the active metal. After molybdopterin is eventually complexed with molybdenum, the complete ligand is usually called molybdenum cofactor.
Bioorganometallic chemistry is the study of biologically active molecules that contain carbon directly bonded to metals or metalloids. The importance of main-group and transition-metal centers has long been recognized as important to the function of enzymes and other biomolecules. However, only a small subset of naturally-occurring metal complexes and synthetically prepared pharmaceuticals are organometallic; that is, they feature a direct covalent bond between the metal(loid) and a carbon atom. The first, and for a long time, the only examples of naturally occurring bioorganometallic compounds were the cobalamin cofactors (vitamin B12) in its various forms. In the 21st century, as a result of the discovery of new systems containing carbon–metal bonds in biology, bioorganometallic chemistry is rapidly emerging as a distinct subdiscipline of bioinorganic chemistry that straddles organometallic chemistry and biochemistry. Naturally occurring bioorganometallics include enzymes and sensor proteins. Also within this realm are synthetically prepared organometallic compounds that serve as new drugs and imaging agents (technetium-99m sestamibi) as well as the principles relevant to the toxicology of organometallic compounds (e.g., methylmercury). Consequently, bioorganometallic chemistry is increasingly relevant to medicine and pharmacology.
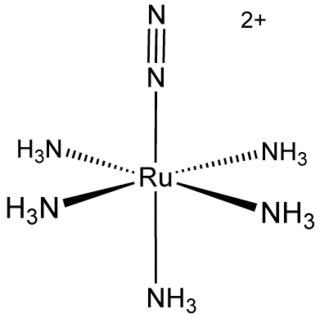
Transition metal dinitrogen complexes are coordination compounds that contain transition metals as ion centers the dinitrogen molecules (N2) as ligands.
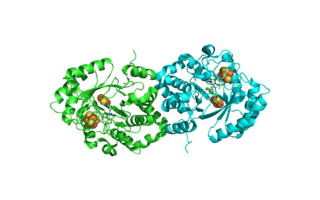
In enzymology, carbon monoxide dehydrogenase (CODH) (EC 1.2.7.4) is an enzyme that catalyzes the chemical reaction
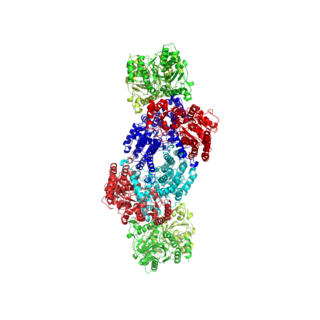
Biotin synthase (BioB) is an enzyme that catalyzes the conversion of dethiobiotin (DTB) to biotin; this is the final step in the biotin biosynthetic pathway. Biotin, also known as vitamin B7, is a cofactor used in carboxylation, decarboxylation, and transcarboxylation reactions in many organisms including humans. Biotin synthase is an S-Adenosylmethionine (SAM) dependent enzyme that employs a radical mechanism to thiolate dethiobiotin, thus converting it to biotin.

Vanadium nitrogenase is a key enzyme for nitrogen fixation found in nitrogen-fixing bacteria, and is used as an alternative to molybdenum nitrogenase when molybdenum is unavailable. Vanadium nitrogenases are an important biological use of vanadium, which is uncommonly used by life. An important component of the nitrogen cycle, vanadium nitrogenase converts nitrogen gas to ammonia, thereby making otherwise inaccessible nitrogen available to plants. Unlike molybdenum nitrogenase, vanadium nitrogenase can also reduce carbon monoxide to ethylene, ethane and propane but both enzymes can reduce protons to hydrogen gas and acetylene to ethylene.

In biochemistry, the iron–sulfur cluster biosynthesis describes the components and processes involved in the biosynthesis of iron–sulfur proteins. The topic is of interest because these proteins are pervasive. The iron sulfur proteins contain iron–sulfur clusters, some with elaborate structures, that feature iron and sulfide centers. One broad biosynthetic task is producing sulfide (S2-), which requires various families of enzymes. Another broad task is affixing the sulfide to iron, which is achieved on scaffolds, which are nonfunctional. Finally these Fe-S cluster is transferred to a target protein, which then become functional.

The Nif regulon is a set of seven operons used to regulate nitrogen fixation in the coliform bacterium Klebsiella pneumoniae under anaerobic and microaerophilic conditions. It includes 17 nif genes, and is situated between the his and the Shi-A operon of the bacterium.
Radical SAMenzymes is a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. Radical SAM enzymes comprise the largest superfamily of metal-containing enzymes.
Evolution of metal ions in biological systems refers to the incorporation of metallic ions into living organisms and how it has changed over time. Metal ions have been associated with biological systems for billions of years, but only in the last century have scientists began to truly appreciate the scale of their influence. Major and minor metal ions have become aligned with living organisms through the interplay of biogeochemical weathering and metabolic pathways involving the products of that weathering. The associated complexes have evolved over time.

Crocosphaera watsonii is an isolate of a species of unicellular diazotrophic marine cyanobacteria which represent less than 0.1% of the marine microbial population. They thrive in offshore, open-ocean oligotrophic regions where the waters are warmer than 24 degrees Celsius. Crocosphaera watsonii cell density can exceed 1,000 cells per milliliter within the euphotic zone; however, their growth may be limited by the concentration of phosphorus. Crocosphaera watsonii are able to contribute to the oceanic carbon and nitrogen budgets in tropical oceans due to their size, abundance, and rapid growth rate. Crocosphaera watsonii are unicellular nitrogen fixers that fix atmospheric nitrogen to ammonia during the night and contribute to new nitrogen in the oceans. They are a major source of nitrogen to open-ocean systems. Nitrogen fixation is important in the oceans as it not only allows phytoplankton to continue growing when nitrogen and ammonium are in very low supply but it also replenishes other forms of nitrogen, thus fertilizing the ocean and allowing more phytoplankton growth.
Serena DeBeer is an American chemist. She is currently a W3-Professor and the director at the Max Planck Institute for Chemical Energy Conversion in Muelheim an der Ruhr, Germany, where she heads the Department of Inorganic Spectroscopy. Her expertise lies in the application and development of X-ray based spectroscopic methods as probes of electronic structure in biological and chemical catalysis.

Metal cluster compounds are a molecular ion or neutral compound composed of three or more metals and featuring significant metal-metal interactions.
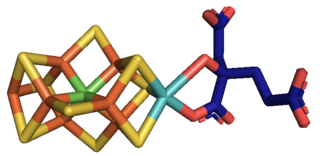
Molybdenum is an essential element in most organisms. It is most notably present in nitrogenase which is an essential part of nitrogen fixation.












